Code Generation for Semantic Segmentation Application on ARM Neon Targets That Uses U-Net
This example shows how to generate code for an image segmentation application that uses deep learning. It uses the codegen command to generate a static library that performs prediction on a DAG Network object for U-Net. U-Net is a deep learning network for image segmentation.
For a similar example that uses U-Net for image segmentation but does not use the codegen command, see Semantic Segmentation of Multispectral Images Using Deep Learning (Image Processing Toolbox).
Prerequisites
ARM® processor that supports the NEON extension and has a RAM of at least 3GB
ARM Compute Library (on the target ARM hardware)
Environment variables for the compilers and libraries
MATLAB® Coder™
MATLAB Coder Interface for Deep Learning support package
Deep Learning Toolbox™
The ARM Compute library version that this example uses might not be the latest version that code generation supports. For information about supported versions of libraries and about environment variables, see Prerequisites for Deep Learning with MATLAB Coder.
This example is not supported in MATLAB Online.
Overview of U-Net
U-Net [1] is a type of convolutional neural network (CNN) designed for semantic image segmentation. In U-Net, the initial series of convolutional layers are interspersed with max pooling layers, successively decreasing the resolution of the input image. These layers are followed by a series of convolutional layers interspersed with upsampling operators, successively increasing the resolution of the input image. The combination of these two series paths forms a U-shaped graph. The U-Net network was originally trained to perform prediction on biomedical image segmentation applications. This example demonstrates the ability of the network to track changes in forest cover over time. Environmental agencies track deforestation to assess and qualify the environmental and ecological health of a region.
Deep learning based semantic segmentation can yield a precise measurement of vegetation cover from high-resolution aerial photographs. One of the challenges of such computation is to differentiating classes that have similar visual characteristics, such as classifying a green pixel as grass, shrubbery, or tree. To increase classification accuracy, some data sets contain multispectral images that provide additional information about each pixel. For example, the Hamlin Beach State Park data set supplements the color images with near-infrared channels that provide a clearer separation of the classes.
This example uses the Hamlin Beach State Park Data [2] along with a pretrained U-Net network to correctly classify each pixel.
The U-Net that this example uses is trained to segment pixels belonging to a set of 18 classes which includes:
0. Other Class/Image Border 7. Picnic Table 14. Grass 1. Road Markings 8. Black Wood Panel 15. Sand 2. Tree 9. White Wood Panel 16. Water (Lake) 3. Building 10. Orange Landing Pad 17. Water (Pond) 4. Vehicle (Car, Truck, or Bus) 11. Water Buoy 18. Asphalt (Parking Lot/Walkway) 5. Person 12. Rocks 6. Lifeguard Chair 13. Other Vegetation
The segmentationUnetARM Entry-Point Function
The segmentationUnetARM entry-point function performs patchwise semantic segmentation on the input image by using the multispectralUnet network contained in the multispectralUnet.mat file. The function loads the network object from the multispectralUnet.mat file into a persistent variable mynet and reuses the persistent variable on subsequent prediction calls.
type('segmentationUnetARM.m')% OUT = segmentationUnetARM(IM) returns a semantically segmented
% image, which is segmented using the network multispectralUnet. This segmentation
% is performed on the input image patchwise on patches of size 256,256.
%
% Copyright 2019-2020 The MathWorks, Inc.
function out = segmentationUnetARM(im)
%#codegen
persistent mynet;
if isempty(mynet)
mynet = coder.loadDeepLearningNetwork('trainedUnet/multispectralUnet.mat');
end
% The input data has to be padded to the size compatible
% with the network Input Size. This input_data is padded inorder to
% perform semantic segmentation on each patch of size (Network Input Size)
[height, width, nChannel] = size(im);
patch = coder.nullcopy(zeros([256, 256, nChannel-1]));
%
padSize = zeros(1,2);
padSize(1) = 256 - mod(height, 256);
padSize(2) = 256 - mod(width, 256);
%
% Pad image must have dimensions as multiples of network input dimensions
im_pad = padarray (im, padSize, 0, 'post');
[height_pad, width_pad, ~] = size(im_pad);
%
out = zeros([size(im_pad,1), size(im_pad,2)], 'uint8');
for i = 1:256:height_pad
for j =1:256:width_pad
for p = 1:nChannel -1
patch(:,:,p) = squeeze( im( i:i+255,...
j:j+255,...
p));
end
% pass in input
segmentedLabels = activations(mynet, patch, 'Segmentation-Layer');
% Takes the max of each channel (6 total at this point)
[~,L] = max(segmentedLabels,[],3);
patch_seg = uint8(L);
% populate section of output
out(i:i+255, j:j+255) = patch_seg;
end
end
% Remove the padding
out = out(1:height, 1:width);
Get Pretrained U-Net DAG Network Object
Download the multispectralUnet.mat file and load the U-Net DAG network object.
if ~exist('trainedUnet/multispectralUnet.mat','file') trainedUnet_url = 'https://www.mathworks.com/supportfiles/vision/data/multispectralUnet.mat'; downloadUNet(trainedUnet_url,pwd); end
ld = load("trainedUnet/multispectralUnet.mat");
net = ld.net;
The DAG network contains 58 layers that include convolution, max pooling, depth concatenation, and pixel classification output layers. To display an interactive visualization of the deep learning network architecture, use the analyzeNetwork (Deep Learning Toolbox) function.
analyzeNetwork(net);
Prepare Input Data
Download the Hamlin Beach State Park data.
if ~exist(fullfile(pwd,'data'),'dir') url = 'http://home.cis.rit.edu/~cnspci/other/data/rit18_data.mat'; downloadHamlinBeachMSIData(url,pwd+"/data/"); end
Load and examine the data in MATLAB.
load(fullfile(pwd,'data','rit18_data','rit18_data.mat'));
Examine data
whos test_data
The image has seven channels. The RGB color channels are the fourth, fifth, and sixth image channels. The first three channels correspond to the near-infrared bands and highlight different components of the image based on their heat signatures. Channel 7 is a mask that indicates the valid segmentation region.
The multispectral image data is arranged as numChannels-by-width-by-height arrays. In MATLAB, multichannel images are arranged as width-by-height-by-numChannels arrays. To reshape the data so that the channels are in the third dimension, use the helper function, switchChannelsToThirdPlane.
test_data = switchChannelsToThirdPlane(test_data);
Confirm data has the correct structure (channels last).
whos test_data
This example uses a cropped version of the full Hamlin Beach State Park dataset that the test_data variable contains. Crop the height and width of test_data to create the variable input_data that this example uses.
test_datacropRGB = imcrop(test_data(:,:,1:3),[2600, 3000, 2000, 2000]); test_datacropInfrared = imcrop(test_data(:,:,4:6),[2600, 3000, 2000, 2000]); test_datacropMask = imcrop(test_data(:,:,7),[2600, 3000, 2000, 2000]);
input_data(:,:,1:3) = test_datacropRGB; input_data(:,:,4:6) = test_datacropInfrared; input_data(:,:,7) = test_datacropMask;
Examine the input_data variable.
whos('input_data');
Write the input data into a text file that is passed as input to the generated executable.
WriteInputDatatoTxt(input_data); [height, width, channels] = size(input_data);
Set Up a Code Generation Configuration Object for a Static Library
To generate code that targets an ARM-based device, create a configuration object for a library. Do not create a configuration object for an executable program. Set up the configuration object for generation of C++ source code only.
cfg = coder.config('lib'); cfg.TargetLang = 'C++'; cfg.GenCodeOnly = true;
Set Up a Configuration Object for Deep Learning Code Generation
Create a coder.ARMNEONConfig object. Specify the library version and the architecture of the target ARM processor. For example, suppose that the target board is a HiKey/Rock960 board with ARMv8 architecture and ARM Compute Library version 20.02.1.
dlcfg = coder.DeepLearningConfig('arm-compute'); dlcfg.ArmComputeVersion = '20.02.1'; dlcfg.ArmArchitecture = 'armv8';
Assign the DeepLearningConfig property of the code generation configuration object cfg to the deep learning configuration object dlcfg.
cfg.DeepLearningConfig = dlcfg;
Generate C++ Source Code by Using codegen
codegen -config cfg segmentationUnetARM -args {ones(size(input_data),'uint16')} -d unet_predict -report
The code gets generated in the unet_predict folder that is located in the current working directory on the host computer.
Generate Zip File by Using packNGo
The packNGo function packages all relevant files into a compressed zip file.
zipFileName = 'unet_predict.zip';
bInfo = load(fullfile('unet_predict','buildInfo.mat'));
packNGo(bInfo.buildInfo, {'fileName', zipFileName,'minimalHeaders', false, 'ignoreFileMissing',true});
The name of the generated zip file is unet_predict.zip.
Copy Generated Zip File to the Target Hardware
Copy the zip file into the target hardware board. Extract the contents of the zip file into a folder and delete the zip file from the hardware.
In the following commands, replace:
passwordwith your passwordusernamewith your user nametargetnamewith the name of your devicetargetDirwith the destination folder for the files
On the Linux® platform, to transfer and extract the zip file on the target hardware, run these commands:
if isunix, system(['sshpass -p password scp -r ' fullfile(pwd,zipFileName) ' username@targetname:targetDir/']), end if isunix, system('sshpass -p password ssh username@targetname "if [ -d targetDir/unet_predict ]; then rm -rf targetDir/unet_predict; fi"'), end if isunix, system(['sshpass -p password ssh username@targetname "unzip targetDir/' zipFileName ' -d targetDir/unet_predict"']), end if isunix, system(['sshpass -p password ssh username@targetname "rm -rf targetDir/' zipFileName '"']), end
On the Windows® platform, to transfer and extract the zip file on the target hardware, run these commands:
if ispc, system(['pscp.exe -pw password -r ' fullfile(pwd,zipFileName) ' username@targetname:targetDir/']), end if ispc, system('plink.exe -l username -pw password targetname "if [ -d targetDir/unet_predict ]; then rm -rf targetDir/unet_predict; fi"'), end if ispc, system(['plink.exe -l username -pw password targetname "unzip targetDir/' zipFileName ' -d targetDir/unet_predict"']), end if ispc, system(['plink.exe -l username -pw password targetname "rm -rf targetDir/' zipFileName '"']), end
Copy Supporting Files to the Target Hardware
Copy these files from the host computer to the target hardware:
Input data,
input_data.txtMakefile for creating the library,
unet_predict_rtw.mkMakefile for building the executable program,
makefile_unet_arm_generic.mk
In the following commands, replace:
passwordwith your passwordusernamewith your user nametargetnamewith the name of your devicetargetDirwith the destination folder for the files
On the Linux® platform, to transfer the supporting files to the target hardware, run these commands:
if isunix, system('sshpass -p password scp unet_predict_rtw.mk username@targetname:targetDir/unet_predict/'), end if isunix, system('sshpass -p password scp input_data.txt username@targetname:targetDir/unet_predict/'), end if isunix, system('sshpass -p password scp makefile_unet_arm_generic.mk username@targetname:targetDir/unet_predict/'), end
On the Windows® platform, to transfer the supporting files to the target hardware, run these commands:
if ispc, system('pscp.exe -pw password unet_predict_rtw.mk username@targetname:targetDir/unet_predict/'), end if ispc, system('pscp.exe -pw password input_data.txt username@targetname:targetDir/unet_predict/'), end if ispc, system('pscp.exe -pw password makefile_unet_arm_generic.mk username@targetname:targetDir/unet_predict/'), end
Build the Library on the Target Hardware
To build the library on the target hardware, execute the generated makefile on the ARM hardware.
Make sure that you set the environment variables ARM_COMPUTELIB and LD_LIBRARY_PATH on the target hardware. See Prerequisites for Deep Learning with MATLAB Coder. The ARM_ARCH variable is used in Makefile to pass compiler flags based on the ARM Architecture. The ARM_VER variable is used in Makefile to compile the code based on the version of the ARM Compute library.
On the Linux host platform, run this command to build the library:
if isunix, system(['sshpass -p password ssh username@targetname "make -C targetDir/unet_predict/ -f unet_predict_rtw.mk ARM_ARCH=' dlcfg.ArmArchitecture ' ARM_VER=' dlcfg.ArmComputeVersion ' "']), end
On the Windows host platform, run this command to build the library:
if ispc, system(['plink.exe -l username -pw password targetname "make -C targetDir/unet_predict/ -f unet_predict_rtw.mk ARM_ARCH=' dlcfg.ArmArchitecture ' ARM_VER=' dlcfg.ArmComputeVersion ' "']), end
Create Executable on the Target
In these commands, replace targetDir with the destination folder where the library is generated. The variables height, width, and channels represent the dimensions of the input data.
main_unet_arm_generic.cpp is the C++ main wrapper file which invokes the segmentationUnetARM function and passes the input image to it. Build the library with the wrapper file to create the executable.
On the Linux host platform, to create the executable, run these commands:
if isunix, system('sshpass -p password scp main_unet_arm_generic.cpp username@targetname:targetDir/unet_predict/'), end if isunix, system(['sshpass -p password ssh username@targetname "make -C targetDir/unet_predict/ IM_H=' num2str(height) ' IM_W=' num2str(width) ' IM_C=' num2str(channels) ' -f makefile_unet_arm_generic.mk"']), end
On the Windows host platform, to create the executable, run these commands:
if ispc, system('pscp.exe -pw password main_unet_arm_generic.cpp username@targetname:targetDir/unet_predict/'), end if ispc, system(['plink.exe -l username -pw password targetname "make -C targetDir/unet_predict/ IM_H=' num2str(height) ' IM_W=' num2str(width) ' IM_C=' num2str(channels) ' -f makefile_unet_arm_generic.mk"']), end
Run the Executable on the Target Hardware
Run the Executable on the target hardware with the input image file input_data.txt.
On the Linux host platform, run this command:
if isunix, system('sshpass -p password ssh username@targetname "cd targetDir/unet_predict/; ./unet input_data.txt output_data.txt"'), end
On the Windows host platform, run this command:
if ispc, system('plink.exe -l username -pw password targetname "cd targetDir/unet_predict/; ./unet input_data.txt output_data.txt"'), end
The unet executable accepts the input data. Because of the large size of input_data (2001x2001x7), it is easier to process the input image in patches. The executable splits the input image into multiple patches, each corresponding to network input size. The executable performs prediction on the pixels in one particular patch at a time and then combines all the patches together.
Transfer the Output from Target Hardware to MATLAB
Copy the generated output file output_data.txt back to the current MATLAB session. On the Linux platform, run:
if isunix, system('sshpass -p password scp username@targetname:targetDir/unet_predict/output_data.txt ./'), end
To perform the same action on the Windows platform, run:
if ispc, system('pscp.exe -pw password username@targetname:targetDir/unet_predict/output_data.txt ./'), end
Store the output data in the variable segmentedImage:
segmentedImage = uint8(importdata('output_data.txt'));
segmentedImage = reshape(segmentedImage,[height,width]);
To extract only the valid portion of the segmented image, multiply it by the mask channel of the input data.
segmentedImage = uint8(input_data(:,:,7)~=0) .* segmentedImage;
Remove the noise and stray pixels by using the medfilt2 function.
segmentedImageCodegen = medfilt2(segmentedImage,[5,5]);
Display U-Net Segmented Data
This line of code creates a vector of the class names.
classNames = net.Layers(end).Classes; disp(classNames);
Overlay the labels on the segmented RGB test image and add a color bar to the segmented image.
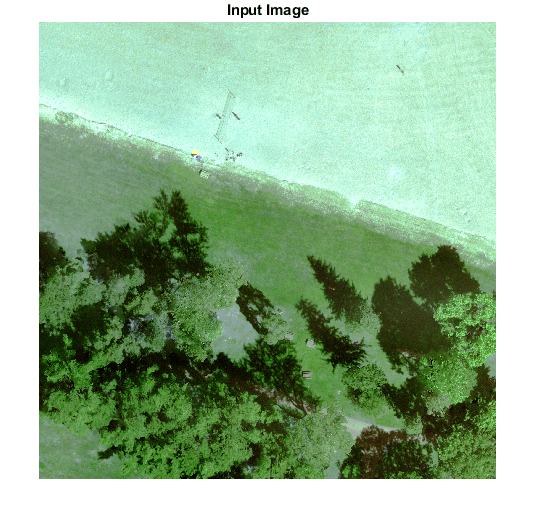
Display input data.
figure(1);
imshow(histeq(input_data(:,:,1:3)));
title('Input Image');
cmap = jet(numel(classNames)); segmentedImageOut = labeloverlay(imadjust(input_data(:,:,4:6),[0 0.6],[0.1 0.9],0.55),segmentedImage,'Transparency',0,'Colormap',cmap); figure(2); imshow(segmentedImageOut);
Display segmented data.
title('Segmented Image using Codegen on ARM'); N = numel(classNames); ticks = 1/(N*2):1/N:1; colorbar('TickLabels',cellstr(classNames),'Ticks',ticks,'TickLength',0,'TickLabelInterpreter','none'); colormap(cmap)
Display segmented overlay image.
segmentedImageOverlay = labeloverlay(imadjust(input_data(:,:,4:6),[0 0.6],[0.1 0.9],0.55),segmentedImage,'Transparency',0.7,'Colormap',cmap); figure(3); imshow(segmentedImageOverlay); title('Segmented Overlay Image');

References
[1] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-Net: Convolutional Networks for Biomedical Image Segmentation." arXiv preprint arXiv:1505.04597, 2015.
[2] Kemker, R., C. Salvaggio, and C. Kanan. "High-Resolution Multispectral Dataset for Semantic Segmentation." CoRR, abs/1703.01918, 2017.
[3] Reference Input Data used is part of the Hamlin Beach State Park data. The following steps can be used to download the data for further evaluation.
if ~exist(fullfile(pwd,'data')) url = 'http://home.cis.rit.edu/~cnspci/other/data/rit18_data.mat'; downloadHamlinBeachMSIData(url,pwd+"/data/"); end
[4] Kemker, Ronald, Carl Salvaggio, and Christopher Kanan. "Algorithms for Semantic Segmentation of Multispectral Remote Sensing Imagery Using Deep Learning." ISPRS Journal of Photogrammetry and Remote Sensing, Deep Learning RS Data, 145 (November 1, 2018): 60-77. https://doi.org/10.1016/j.isprsjprs.2018.04.014.
See Also
coder.ARMNEONConfig | coder.DeepLearningConfig | coder.hardware | packNGo