Test Metrics in Modelscape
This example shows how to implement test metrics in MATLAB® using Modelscape™ software.
For information about test metrics from the model development and validation point of view, see Credit Scorecard Validation Metrics and Fairness Metrics in Modelscape, respectively.
Write Test Metrics
The basic building block of Modelscape metrics framework is the mrm.data.validation.TestMetric class. This class defines these properties:
Name— Name for the test metric, specified as a string scalar.ShortName— Concise name for accessing metrics inMetricsHandlerobjects, specified as a valid MATLAB property name.Value— Values that the metric carries, specified as a numeric scalar or row vector.Keys— Value parameterization keys, specified as an -by- string array. is the length ofValue. The keys default to an empty string.KeyNames— Key names, specified as a length- string vector, where is the length ofValue. The default value is"Key".Diagnostics— Diagnostics related to metric calculation, specified as a structure.
Any subclass of TestMetric must implement a constructor and a compute method to fill in these values.
For example, the Modelscape statistical parity difference (SPD) metric for bias detection has Name "Statistical Parity Difference" and a ShortName "StatisticalParityDifference". This table shows the arrangement of Keys and KeyNames.
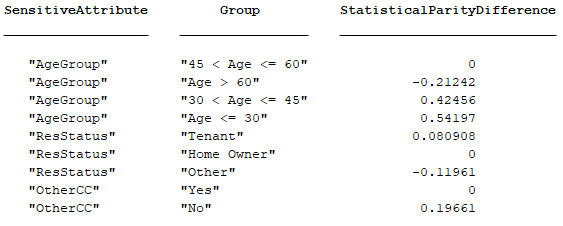
The KeyNames property comprises the values "SensitiveAttribute" and "Group". The Keys property comprises the two columns with attribute-group combinations. The ShortName property value appears as the third header. The third column of the table carries the Value property value of the metric.
The base class has these methods, which you can override:
ComparisonValue(this)— Change the value against which the software compares the threshold. For example, in statistical hypothesis testing, this method returns the p-value of the computed statistic.formatResult(this)— Table for the SPD metric.project(this)— Restriction of a non-scalar metric to a subset of keys. Extend the default implementation in a subclass to cover any diagnostic or auxiliary data carried that the subclass objects carry.
Write Metrics With Visualizations
To write test metrics equipped with visualizations, define metrics that inherit from mrm.data.validation.TestMetricWithVisualization. This class adds an additional requirement to the TestMetric base class to implement a visualization method with the signature fig = visualize(this, options). Use options to define name-value arguments for the metric. For example, use a sensitive attribute with the StatisticalParityDifference metric for visualization.
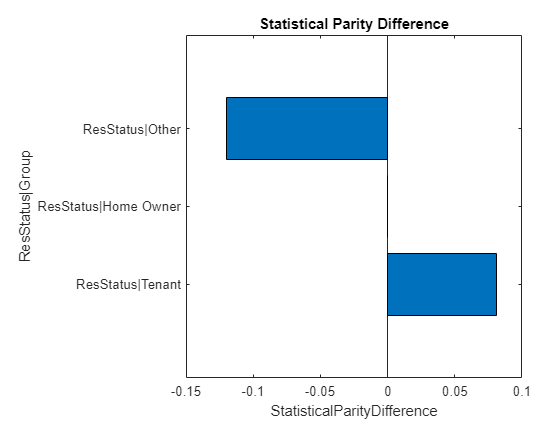
spdFig = visualize(spdMetric,SensitiveAttribute="ResStatus");
Write Metrics Projecting onto Selected Keys
The visualization shows the SPD metrics for the ResStatus attribute only. This plot uses the project method of the TestMetric class which uses selected keys of a metric. For a metric with key names, project accepts an array of up to strings as the Keys property value. The output restricts the metric to keys for which the th element matches the th element of the array.
spdResStatus = project(spdMetric,Keys="ResStatus")

When you specify both keys, the result is a scalar metric:
spdTenant = project(spdMetric,Keys=["ResStatus","Tenant"])
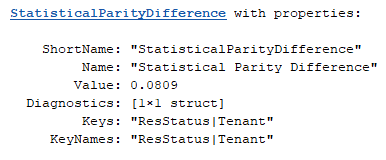
The base class implementation of project does not handle diagnostics or other auxiliary data that the subclass carries. If necessary, implement diagnostics and auxiliary data handling in the subclass using the secondary keySelection output in project.
Write Summarizable Metrics
Summary metrics reveal a different aspect of nonscalar metrics. In the case of the SPD metric, across all the attribute-group pairs, the summary SPD value is the value with the largest deviation from the completely unbiased value of zero.
spdSummary = summary(spdMetric)

Summarize a TestMetric class by inheriting from the mrm.data.validation.TestMetricWithSummaryValue class and implementing the abstract summary method. This method returns a metric of the same type with a singleton Value property. The meaning of the summary value depends on the metric, so this method has no default implementation. However, you can use the protected summaryCore method in TestMetricWithSummaryValue.
Write Test Thresholds
Test metrics are often compared against thresholds to qualitatively assess the inputs. For example, for certain models a model validator can require an area under the ROC curve (AUROC) of at least 0.8 for an acceptable model. Values under 0.7 are red flags, and values between 0.7 and 0.8 require a closer look.
Use the mrm.data.validation.TestThresholds class to implement these thresholds. Encode the thresholds and classifications into a TestThresholds object.
aurocThresholds = mrm.data.validation.TestThresholds([0.7, 0.8], ["Fail", "Undecided", "Pass"]);
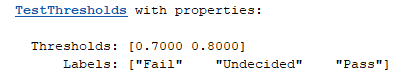
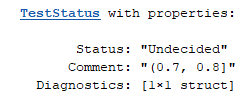
These thresholds and labels govern the output of the status method of TestThresholds. For example, call status with an AUROC threshold of 0.72.
status(aurocThresholds, 0.72)
returns the following. Comment indicates the interval to which the given input belongs.
Customize Thresholds
Implement thresholding regimes or different diagnostics as subclasses of mrm.data.validation.TestThresholdsBase. Implement the status method of the class to populate the Comment and Diagnostics properties according to your needs.
Write Statistical Hypothesis Tests
In some cases, notably in statistical hypothesis testing, the relevant quantity to compare against test thresholds is the associated p-value under a relevant null hypothesis. In these cases, use the test metric class to override the ComparisonValue method and return the p-value instead of the Value property of the metric. For an example, see the Modelscape implementation of the Augmented Dickey-Fuller test.
edit mrm.data.validation.timeseries.ADFMetric
Set the thresholds against which to compare the p-values. This TestThresholds object returns status as "Reject" for p-values less than 0.05 and "Accept" otherwise.
adfThreshold = mrm.data.validation.PValueThreshold(0.05)
