tsne Settings
This example shows the effects of various tsne settings.
Load and Examine Data
Load the humanactivity data, which is available when you run this example.
load humanactivityView a description of the data.
Description
Description = 29×1 string
" === Human Activity Data === "
" "
" The humanactivity data set contains 24,075 observations of five different "
" physical human activities: Sitting, Standing, Walking, Running, and "
" Dancing. Each observation has 60 features extracted from acceleration "
" data measured by smartphone accelerometer sensors. The data set contains "
" the following variables: "
" "
" * actid - Response vector containing the activity IDs in integers: 1, 2, "
" 3, 4, and 5 representing Sitting, Standing, Walking, Running, and "
" Dancing, respectively "
" * actnames - Activity names corresponding to the integer activity IDs "
" * feat - Feature matrix of 60 features for 24,075 observations "
" * featlabels - Labels of the 60 features "
" "
" The Sensor HAR (human activity recognition) App [1] was used to create "
" the humanactivity data set. When measuring the raw acceleration data with "
" this app, a person placed a smartphone in a pocket so that the smartphone "
" was upside down and the screen faced toward the person. The software then "
" calibrated the measured raw data accordingly and extracted the 60 "
" features from the calibrated data. For details about the calibration and "
" feature extraction, see [2] and [3], respectively. "
" "
" [1] El Helou, A. Sensor HAR recognition App. MathWorks File Exchange "
" http://www.mathworks.com/matlabcentral/fileexchange/54138-sensor-har-recognition-app "
" [2] STMicroelectronics, AN4508 Application note. “Parameters and "
" calibration of a low-g 3-axis accelerometer.” 2014. "
" [3] El Helou, A. Sensor Data Analytics. MathWorks File Exchange "
" https://www.mathworks.com/matlabcentral/fileexchange/54139-sensor-data-analytics--french-webinar-code- "
The data set is organized by activity type. To better represent a random set of data, shuffle the rows.
n = numel(actid); % Number of data points rng default % For reproducibility idx = randsample(n,n); % Shuffle X = feat(idx,:); % Shuffled data actid = actid(idx); % Shuffled labels
Associate the activities with the labels in actid.
activities = ["Sitting";"Standing";"Walking";"Running";"Dancing"]; activity = activities(actid);
Process Data Using t-SNE
Obtain two-dimensional analogs of the data clusters using t-SNE.
rng default % For reproducibility Y = tsne(X); figure numGroups = length(unique(actid)); clr = hsv(numGroups); gscatter(Y(:,1),Y(:,2),activity,clr) title('Default Figure')
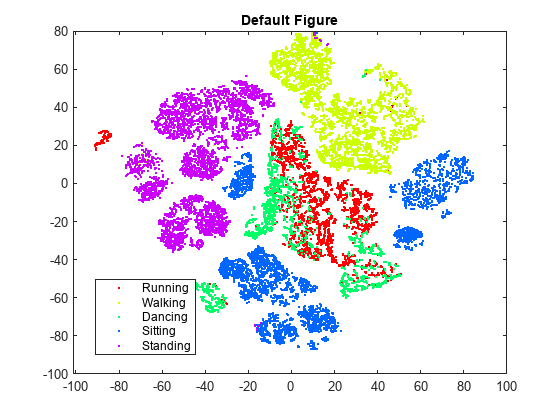
t-SNE creates a figure with relatively few data points that seem misplaced. However, the clusters are not very well separated.
Perplexity
Try altering the perplexity setting to see the effect on the figure.
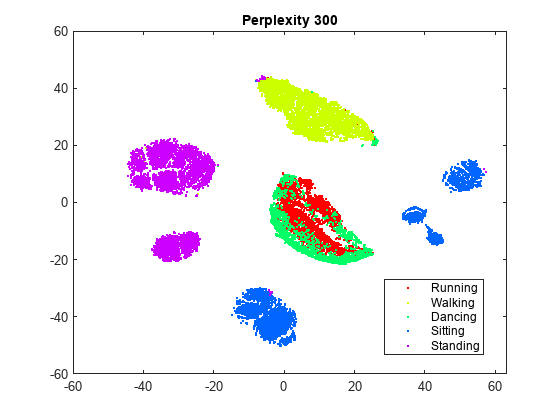
rng default % For fair comparison Y300 = tsne(X,Perplexity=300); figure gscatter(Y300(:,1),Y300(:,2),activity,clr) title('Perplexity 300')
rng default % For fair comparison Y4 = tsne(X,Perplexity=4); figure gscatter(Y4(:,1),Y4(:,2),activity,clr) title('Perplexity 4')

Setting the perplexity to 300 gives a figure that has better-separated clusters than the original figure. Setting the perplexity to 4 gives a figure without well separated clusters. For the remainder of this example, use a perplexity of 300.
Exaggeration
Try altering the exaggeration setting to see the effect on the figure.
rng default % For fair comparison YEX20 = tsne(X,Perplexity=300,Exaggeration=20); figure gscatter(YEX20(:,1),YEX20(:,2),activity,clr) title('Exaggeration 20')

rng default % For fair comparison YEx15 = tsne(X,Perplexity=300,Exaggeration=1.5); figure gscatter(YEx15(:,1),YEx15(:,2),activity,clr) title('Exaggeration 1.5')
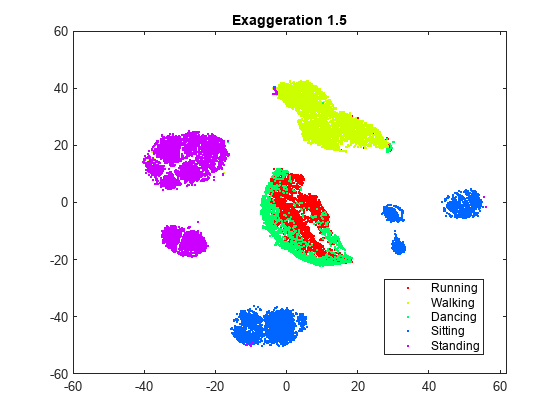
While the exaggeration setting has an effect on the figure, it is not clear whether any nondefault setting gives a better picture than the default setting. The figure with an exaggeration of 20 is similar to the default figure, except the clusters are not quite as well separated. In general, a larger exaggeration creates more empty space between embedded clusters. An exaggeration of 1.5 gives a figure similar to the default exaggeration. Exaggerating the values in the joint distribution of X makes the values in the joint distribution of Y smaller. This makes it much easier for the embedded points to move relative to one another.
Learning Rate
Try altering the learning rate setting to see the effect on the figure.
rng default % For fair comparison YL5 = tsne(X,Perplexity=300,LearnRate=5); figure gscatter(YL5(:,1),YL5(:,2),activity,clr) title('Learning Rate 5')

rng default % For fair comparison YL2000 = tsne(X,Perplexity=300,LearnRate=2000); figure gscatter(YL2000(:,1),YL2000(:,2),activity,clr) title('Learning Rate 2000')

The figure with a learning rate of 5 has several clusters that split into two or more pieces. This shows that if the learning rate is too small, the minimization process can get stuck in a bad local minimum. A learning rate of 2000 gives a figure similar to the figure with no setting of the learning rate.
Initial Behavior with Various Settings
Large learning rates or large exaggeration values can lead to undesirable initial behavior. To see this, set large values of these parameters and set NumPrint and Verbose to 1 to show all the iterations. Stop the iterations after 10, as the goal of this experiment is simply to look at the initial behavior.
Begin by setting the exaggeration to 200.
rng default % For fair comparison opts = statset(MaxIter=10); YEX5000 = tsne(X,Perplexity=300,Exaggeration=5000,... NumPrint=1,Verbose=1,Options=opts);
|==============================================| | ITER | KL DIVERGENCE | NORM GRAD USING | | | FUN VALUE USING | EXAGGERATED DIST| | | EXAGGERATED DIST| OF X | | | OF X | | |==============================================| | 1 | 6.388137e+04 | 6.483115e-04 | | 2 | 6.388775e+04 | 5.267770e-01 | | 3 | 7.131506e+04 | 5.754291e-02 | | 4 | 7.234772e+04 | 6.705418e-02 | | 5 | 7.409144e+04 | 9.278330e-02 | | 6 | 7.484659e+04 | 1.022587e-01 | | 7 | 7.445701e+04 | 9.934864e-02 | | 8 | 7.391345e+04 | 9.633570e-02 | | 9 | 7.315999e+04 | 1.027610e-01 | | 10 | 7.265936e+04 | 1.033174e-01 |
The Kullback-Leibler divergence increases during the first few iterations, and the norm of the gradient increases as well.
To see the final result of the embedding, allow the algorithm to run to completion using the default stopping criteria.
rng default % For fair comparison YEX5000 = tsne(X,Perplexity=300,Exaggeration=5000); figure gscatter(YEX5000(:,1),YEX5000(:,2),activity,clr) title('Exaggeration 5000')

This exaggeration value does not give a clean separation into clusters.
Show the initial behavior when the learning rate is 1,000,000.
rng default % For fair comparison YL1000k = tsne(X,Perplexity=300,LearnRate=1e6,... NumPrint=1,Verbose=1,Options=opts);
|==============================================| | ITER | KL DIVERGENCE | NORM GRAD USING | | | FUN VALUE USING | EXAGGERATED DIST| | | EXAGGERATED DIST| OF X | | | OF X | | |==============================================| | 1 | 2.258150e+01 | 4.412730e-07 | | 2 | 2.259045e+01 | 4.857725e-04 | | 3 | 2.945552e+01 | 3.210405e-05 | | 4 | 2.976546e+01 | 4.337510e-05 | | 5 | 2.976928e+01 | 4.626810e-05 | | 6 | 2.969205e+01 | 3.907617e-05 | | 7 | 2.963695e+01 | 4.943976e-05 | | 8 | 2.960336e+01 | 4.572338e-05 | | 9 | 2.956194e+01 | 6.208571e-05 | | 10 | 2.952132e+01 | 5.253798e-05 |
Again, the Kullback-Leibler divergence increases during the first few iterations, and the norm of the gradient increases as well.
To see the final result of the embedding, allow the algorithm to run to completion using the default stopping criteria.
rng default % For fair comparison YL1000k = tsne(X,Perplexity=300,LearnRate=1e6); figure gscatter(YL1000k(:,1),YL1000k(:,2),activity,clr) title('Learning Rate 1,000,000')
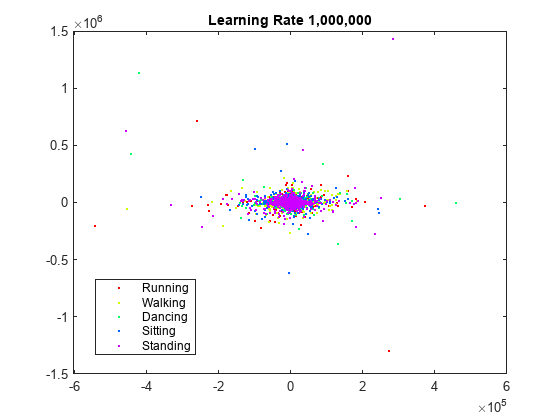
The learning rate is far too large, and gives no useful embedding.
Conclusion
tsne with default settings does a good job of embedding the high-dimensional initial data into two-dimensional points that have well defined clusters. Increasing the perplexity gives a better-looking embedding with this data. The effects of algorithm settings are difficult to predict. Sometimes they can improve the clustering, but for typically the default settings seem good. While speed is not part of this investigation, settings can affect the speed of the algorithm. In particular, the default Barnes-Hut algorithm is notably faster on this data.