codonbias
Calculate codon frequency for each amino acid coded for in nucleotide sequence
Syntax
CodonFreq = codonbias(SeqNT)
CodonFreq = codonbias(SeqNT,
...'GeneticCode', GeneticCodeValue, ...)
CodonFreq = codonbias(SeqNT,
...'Frame', FrameValue, ...)
CodonFreq = codonbias(SeqNT,
...'Reverse', ReverseValue, ...)
CodonFreq = codonbias(SeqNT,
...'Ambiguous', AmbiguousValue, ...)
CodonFreq = codonbias(SeqNT,
...'Pie', PieValue, ...)
Input Arguments
SeqNT | One of the following:
Valid characters include
|
GeneticCodeValue | Integer, character vector, or string specifying a genetic code
number or code name from the table Genetic Code. Default is
Tip If you use a code name, you can truncate the name to the first two letters of the name. |
FrameValue | Integer specifying a reading frame
in the nucleotide sequence. Choices are |
ReverseValue | Controls the return of the codon frequency
for the reverse complement sequence of the nucleotide sequence specified
by SeqNT. Choices are true or false (default). |
AmbiguousValue | Character vector or string specifying how to treat codons containing ambiguous
nucleotide characters (
|
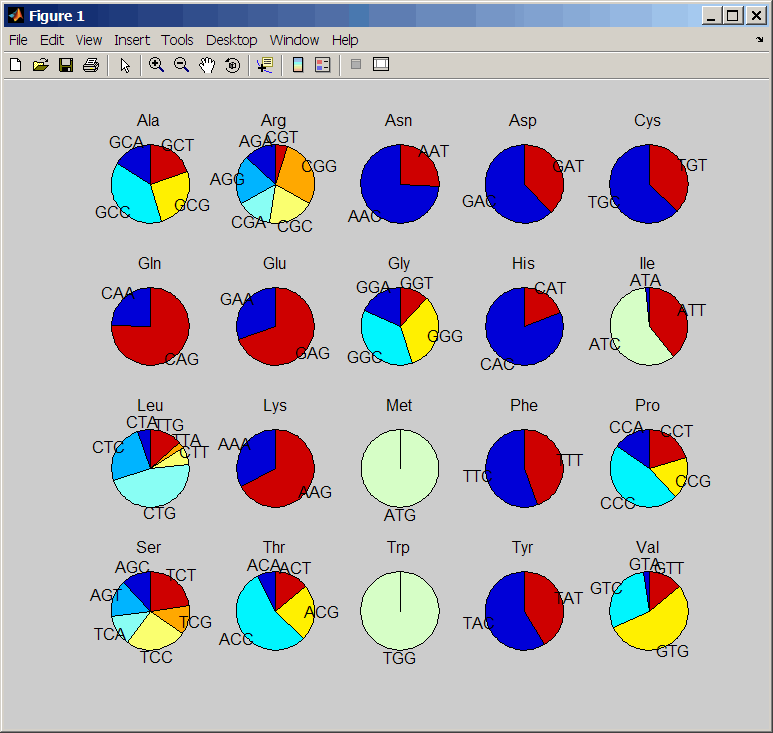
PieValue | Controls the creation of a figure of
20 pie charts, one for each amino acid. Choices are true or false (default). |
Output Arguments
CodonFreq | MATLAB structure containing a field for each amino acid, each of which contains the associated codon frequencies as percentages. |
Description
Many amino acids are coded by two or more nucleic acid codons. However, the probability that a specific codon (from all possible codons for an amino acid) is used to code an amino acid varies between sequences. Knowing the frequency of each codon in a protein coding sequence for each amino acid is a useful statistic.
CodonFreq = codonbias(SeqNT)SeqNT,
a nucleotide sequence, and returns the results in CodonFreq,
a MATLAB structure containing a field for each amino acid.
CodonFreq = codonbias(SeqNT,
...'PropertyName', PropertyValue,
...)codonbias with optional properties
that use property name/property value pairs. You can specify one or
more properties in any order. Each PropertyName must
be enclosed in single quotation marks and is case insensitive. These
property name/property value pairs are as follows:
specifies a genetic code. Choices for CodonFreq = codonbias(SeqNT,
...'GeneticCode', GeneticCodeValue, ...)GeneticCodeValue are an
integer, character vector, or string specifying a code number or code name from the
table Genetic Code. If you use a code name, you can truncate the name to the
first two characters of the name. Default is 1 or
'Standard'.
Tip
If you use a code name, you can truncate the name to the first two letters of the name.
CodonFreq = codonbias(SeqNT,
...'Frame', FrameValue, ...)FrameValue,
which can be 1 (default), 2,
or 3.
CodonFreq = codonbias(SeqNT,
...'Reverse', ReverseValue, ...)SeqNT.
Choices are true or false (default).
CodonFreq = codonbias(SeqNT,
...'Ambiguous', AmbiguousValue, ...)'ignore' (default), 'prorate',
and 'warn'.
CodonFreq = codonbias(SeqNT,
...'Pie', PieValue, ...)true or false (default).
Genetic Code
| Code Number | Code Name |
|---|---|
1 | Standard |
2 | Vertebrate Mitochondrial |
3 | Yeast Mitochondrial |
4 | Mold, Protozoan, Coelenterate
Mitochondrial, and Mycoplasma/Spiroplasma |
5 | Invertebrate Mitochondrial |
6 | Ciliate, Dasycladacean,
and Hexamita Nuclear |
9 | Echinoderm Mitochondrial |
10 | Euplotid Nuclear |
11 | Bacterial and Plant Plastid |
12 | Alternative Yeast Nuclear |
13 | Ascidian Mitochondrial |
14 | Flatworm Mitochondrial |
15 | Blepharisma Nuclear |
16 | Chlorophycean Mitochondrial |
21 | Trematode Mitochondrial |
22 | Scenedesmus Obliquus Mitochondrial |
23 | Thraustochytrium Mitochondrial |
Examples
Version History
Introduced before R2006a