fitrqlinear
Syntax
Description
Mdl = fitrqlinear(Tbl,ResponseVarName)Mdl. The function
trains the model using the predictors in the table Tbl and the response
values in the ResponseVarName table variable.
By default, the function uses the median (0.5 quantile).
Mdl = fitrqlinear(___,Name=Value)Quantiles name-value argument.
[
also returns Mdl,AggregateOptimizationResults] = fitrqlinear(___)AggregateOptimizationResults, which contains
hyperparameter optimization results when you specify the
OptimizeHyperparameters and
HyperparameterOptimizationOptions name-value arguments. You must also
specify the ConstraintType and ConstraintBounds
options of HyperparameterOptimizationOptions. You can use this syntax
to optimize on the compact model size instead of the cross-validation loss, and to solve a
set of multiple optimization problems that have the same options but different constraint
bounds. (since R2025a)
Note
Hyperparameter optimization is supported only for models with one quantile.
Examples
Fit a quantile linear regression model using the 0.25, 0.50, and 0.75 quantiles.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s. Create a matrix X containing the predictor variables Acceleration, Displacement, Horsepower, and Weight. Store the response variable MPG in the variable Y.
load carbig
X = [Acceleration,Displacement,Horsepower,Weight];
Y = MPG;Delete rows of X and Y where either array has missing values.
R = rmmissing([X Y]); X = R(:,1:end-1); Y = R(:,end);
Partition the data into training data (XTrain and YTrain) and test data (XTest and YTest). Reserve approximately 20% of the observations for testing, and use the rest of the observations for training.
rng(0,"twister") % For reproducibility of the partition c = cvpartition(length(Y),"Holdout",0.20); trainingIdx = training(c); XTrain = X(trainingIdx,:); YTrain = Y(trainingIdx); testIdx = test(c); XTest = X(testIdx,:); YTest = Y(testIdx);
Train a quantile linear regression model. Specify to use the 0.25, 0.50, and 0.75 quantiles (that is, the lower quartile, median, and upper quartile). To improve the model fit, change the beta tolerance to 1e-6 instead of the default value 1e-4. Use a ridge (L2) regularization term of 1. Adjusting the regularization term can help prevent quantile crossing.
Mdl = fitrqlinear(XTrain,YTrain,Quantiles=[0.25,0.50,0.75], ...
BetaTolerance=1e-6,Lambda=1)Mdl =
RegressionQuantileLinear
ResponseName: 'Y'
CategoricalPredictors: []
ResponseTransform: 'none'
Beta: [4×3 double]
Bias: [17.0004 23.0029 29.5243]
Quantiles: [0.2500 0.5000 0.7500]
Properties, Methods
Mdl is a RegressionQuantileLinear model object. You can use dot notation to access the properties of Mdl. For example, Mdl.Beta and Mdl.Bias contain the linear coefficient estimates and estimated bias terms, respectively. Each column of Mdl.Beta corresponds to one quantile, as does each element of Mdl.Bias.
In this example, you can use the linear coefficient estimates and estimated bias terms directly to predict the test set responses for each of the three quantiles in Mdl.Quantiles. In general, you can use the predict object function to make quantile predictions.
predictedY = XTest*Mdl.Beta + Mdl.Bias
predictedY = 78×3
12.3963 16.2569 19.5263
5.8328 10.1568 12.6058
17.1726 20.6398 24.9748
23.3790 28.1122 31.3617
17.0036 22.5314 23.0539
16.6120 17.0713 20.1062
10.9274 12.3302 13.2707
14.9130 14.6659 12.7100
16.3103 17.7497 20.8477
19.6229 25.7109 30.5389
19.5583 24.6621 30.4345
12.9525 14.4508 16.0004
14.8525 16.1338 16.4112
24.1648 31.1758 33.9310
15.1039 17.8497 19.2013
⋮
isequal(predictedY,predict(Mdl,XTest))
ans = logical
1
Each column of predictedY corresponds to a separate quantile (0.25, 0.5, or 0.75).
Visualize the predictions of the quantile linear regression model. First, create a grid of predictor values.
minX = floor(min(X))
minX = 1×4
8 68 46 1613
maxX = ceil(max(X))
maxX = 1×4
25 455 230 5140
gridX = zeros(100,size(X,2)); for p = 1:size(X,2) gridp = linspace(minX(p),maxX(p))'; gridX(:,p) = gridp; end
Next, use the trained model Mdl to predict the response values for the grid of predictor values.
gridY = predict(Mdl,gridX)
gridY = 100×3
20.8073 25.4104 29.1436
20.6991 25.2907 29.0251
20.5909 25.1711 28.9066
20.4828 25.0514 28.7881
20.3746 24.9318 28.6696
20.2664 24.8121 28.5512
20.1583 24.6924 28.4327
20.0501 24.5728 28.3142
19.9419 24.4531 28.1957
19.8337 24.3335 28.0772
19.7256 24.2138 27.9587
19.6174 24.0941 27.8402
19.5092 23.9745 27.7217
19.4011 23.8548 27.6032
19.2929 23.7351 27.4848
⋮
For each observation in gridX, the predict object function returns predictions for the quantiles in Mdl.Quantiles.
View the gridY predictions for the second predictor (Displacement). Compare the quantile predictions to the true test data values.
predictorIdx = 2; plot(XTest(:,predictorIdx),YTest,".") hold on plot(gridX(:,predictorIdx),gridY(:,1)) plot(gridX(:,predictorIdx),gridY(:,2)) plot(gridX(:,predictorIdx),gridY(:,3)) hold off xlabel("Predictor (Displacement)") ylabel("Response (MPG)") legend(["True values","0.25 predicted values", ... "0.50 predicted values","0.75 predicted values"]) title("Test Data")
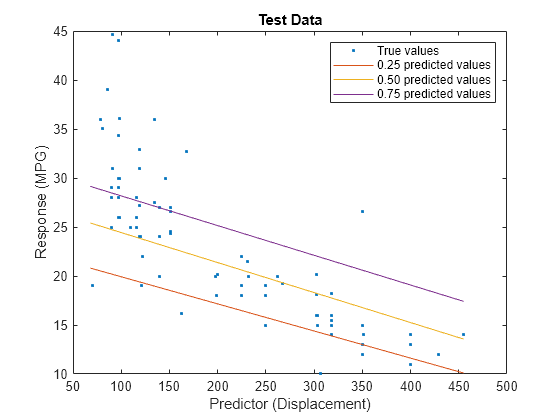
The red line shows the predictions for the 0.25 quantile, the yellow line shows the predictions for the 0.50 quantile, and the purple line shows the predictions for the 0.75 quantile. The blue points indicate the true test data values.
Notice that the quantile prediction lines do not cross each other.
When training a quantile linear regression model, you can use a ridge (L2) regularization term to prevent quantile crossing.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s. Create a table containing the predictor variables Acceleration, Cylinders, Displacement, and so on, as well as the response variable MPG.
load carbig cars = table(Acceleration,Cylinders,Displacement, ... Horsepower,Model_Year,Weight,MPG);
Remove rows of cars where the table has missing values.
cars = rmmissing(cars);
Partition the data into training and test sets using cvpartition. Use approximately 80% of the observations as training data, and 20% of the observations as test data.
rng(0,"twister") % For reproducibility of the data partition c = cvpartition(height(cars),"Holdout",0.20); trainingIdx = training(c); carsTrain = cars(trainingIdx,:); testIdx = test(c); carsTest = cars(testIdx,:);
Train a quantile linear regression model. Use the 0.25, 0.50, and 0.75 quantiles (that is, the lower quartile, median, and upper quartile). To improve the model fit, change the beta tolerance to 1e-6 instead of the default value 1e-4.
Mdl = fitrqlinear(carsTrain,"MPG",Quantiles=[0.25 0.5 0.75], ... BetaTolerance=1e-6);
Mdl is a RegressionQuantileLinear model object.
Determine if the test data predictions for the quantiles in Mdl.Quantiles cross each other by using the predict object function of Mdl. The crossingIndicator output argument contains a value of 1 (true) for any observation with quantile predictions that cross.
[~,crossingIndicator] = predict(Mdl,carsTest); sum(crossingIndicator)
ans = 2
In this example, two of the observations in carsTest have quantile predictions that cross each other.
To prevent quantile crossing, specify the Lambda name-value argument in the call to fitrqlinear. Use a 0.1 ridge (L2) penalty term.
newMdl = fitrqlinear(carsTrain,"MPG",Quantiles=[0.25 0.5 0.75], ... BetaTolerance=1e-6,Lambda=0.1); [predictedY,newCrossingIndicator] = predict(newMdl,carsTest); sum(newCrossingIndicator)
ans = 0
With regularization, the predictions for the test data set do not cross for any observations.
Visualize the predictions returned by newMdl by using a scatter plot with a reference line. Plot the predicted values along the vertical axis and the true response values along the horizontal axis. Points on the reference line indicate correct predictions.
plot(carsTest.MPG,predictedY(:,1),".") hold on plot(carsTest.MPG,predictedY(:,2),".") plot(carsTest.MPG,predictedY(:,3),".") plot(carsTest.MPG,carsTest.MPG) hold off xlabel("True MPG") ylabel("Predicted MPG") legend(["0.25 quantile values","0.50 quantile values", ... "0.75 quantile values","Reference line"], ... Location="southeast") title("Test Data")
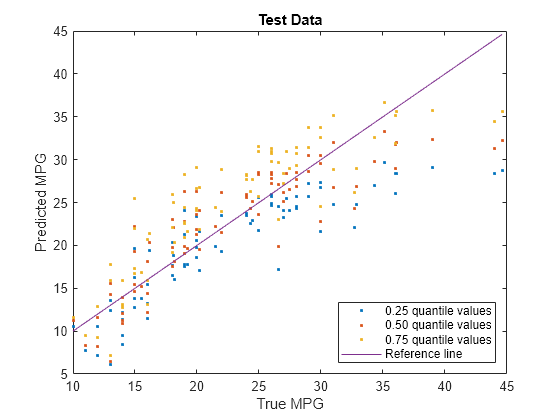
Blue points correspond to the 0.25 quantile, red points correspond to the 0.50 quantile, and yellow points correspond to the 0.75 quantile.
For a more in-depth example, see Regularize Quantile Regression Model to Prevent Quantile Crossing.
Input Arguments
Sample data used to train the model, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one predictor variable.
Optionally, Tbl can contain one additional column for the response
variable. Multicolumn variables and cell arrays other than cell arrays of character
vectors are not allowed.
If
Tblcontains the response variable, and you want to use all remaining variables inTblas predictors, then specify the response variable by usingResponseVarName.If
Tblcontains the response variable, and you want to use only a subset of the remaining variables inTblas predictors, then specify a formula by usingformula.If
Tbldoes not contain the response variable, then specify a response variable by usingY. The length of the response variable and the number of rows inTblmust be equal.
Response variable name, specified as the name of a variable in
Tbl. The response variable must be a numeric vector.
You must specify ResponseVarName as a character vector or string
scalar. For example, if Tbl stores the response variable
Y as Tbl.Y, then specify it as
"Y". Otherwise, the software treats all columns of
Tbl, including Y, as predictors when
training the model.
Data Types: char | string
Explanatory model of the response variable and a subset of the predictor variables,
specified as a character vector or string scalar in the form
"Y~x1+x2+x3". In this form, Y represents the
response variable, and x1, x2, and
x3 represent the predictor variables.
To specify a subset of variables in Tbl as predictors for
training the model, use a formula. If you specify a formula, then the software does not
use any variables in Tbl that do not appear in
formula.
The variable names in the formula must be both variable names in Tbl
(Tbl.Properties.VariableNames) and valid MATLAB® identifiers. You can verify the variable names in Tbl by
using the isvarname function. If the variable names
are not valid, then you can convert them by using the matlab.lang.makeValidName function.
Data Types: char | string
Predictor data used to train the model, specified as a numeric matrix.
By default, the software treats each row of X as one
observation, and each column as one predictor.
The length of Y and the number of observations in
X must be equal.
To specify the names of the predictors in the order of their appearance in
X, use the PredictorNames name-value
argument.
Note
If you orient your predictor matrix so that observations correspond to columns and
specify ObservationsIn="columns", then you might experience a
significant reduction in computation time.
Data Types: single | double
Note
The software treats NaN, empty character vector
(''), empty string (""),
<missing>, and <undefined> elements as
missing values, and removes observations with any of these characteristics:
Missing value in the response
At least one missing value in a predictor observation
NaNvalue or0weight
For economical memory usage, a best practice is to manually remove training observations
that contain missing values before passing the data to
fitrqlinear.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: fitrqlinear(Tbl,"MPG",Quantiles=[0.25 0.5
0.75],Standardize=true) specifies to use the 0.25, 0.5, and 0.75 quantiles and
to standardize the data before training.
Linear Regression Options
Quantiles to use for training Mdl, specified as a vector of
values in the range [0,1]. For each quantile q, the function fits a
linear regression model that separates the bottom 100*q percent of training responses from the top 100*(1 – q) percent of training responses.
You can find the estimated linear model coefficients and estimated bias term for
each quantile in the Beta and Bias properties of Mdl, respectively.
Example: Quantiles=[0.25 0.5 0.75]
Data Types: single | double
Initial coefficient estimates, specified as a
p-by-q numeric matrix. p is
the number of predictor variables after dummy variables are created for categorical
variables (for more details, see CategoricalPredictors), and
q is the number of quantiles (for more details, see
Quantiles).
By default, Beta is a matrix of 0
values.
Data Types: single | double
Initial intercept estimates, specified as a numeric vector of length
q, where q is the number of quantiles (for
more details, see Quantiles).
By default, the initial bias for each quantile is the corresponding weighted quantile of the response.
Data Types: single | double
Flag to include the linear model intercept, specified as true
or false.
| Value | Description |
|---|---|
true | For each quantile, the software includes the bias term b in the linear model, and then estimates it. |
false | The software sets b = 0 during estimation. |
Example: FitBias=false
Data Types: logical
Predictor data observation dimension, specified as "rows" or
"columns".
Note
If you orient your predictor matrix so that observations correspond to columns and
specify ObservationsIn="columns", then you might experience a
significant reduction in computation time. You cannot specify
ObservationsIn="columns" for predictor data in a
table.
Example: ObservationsIn="columns"
Data Types: char | string
Flag to standardize the predictor data, specified as a numeric or logical
0 (false) or 1
(true). If you set Standardize to
true, then the software centers and scales each numeric predictor
variable by the corresponding column mean and standard deviation. The software does not
standardize categorical predictors.
Example:
Standardize=true
Data Types: single | double | logical
Convergence Control Options
Verbosity level, specified as a nonnegative integer. Verbose
controls the amount of diagnostic information fitrqlinear
displays at the command line.
| Value | Description |
|---|---|
0 | fitrqlinear does not display diagnostic
information. |
1 | fitrqlinear periodically displays and stores the
value of the objective function, gradient magnitude, and other diagnostic
information. |
| Any other positive integer | fitrqlinear displays and stores diagnostic
information at each training process iteration. |
Example: Verbose=1
Data Types: single | double
Relative tolerance on the linear coefficients and the bias term (intercept) for each quantile, specified as a nonnegative scalar.
Let , that is, the vector of the coefficients and the bias term at iteration t of the training process. If , then the training process terminates.
Example: BetaTolerance=1e-6
Data Types: single | double
Absolute gradient tolerance for each quantile, specified as a nonnegative scalar.
Let be the gradient vector of the objective function with respect to the coefficients and bias term at iteration t of the training process. If , then the training process terminates.
If you also specify BetaTolerance, then the training process
terminates when fitrqlinear satisfies either stopping
criterion.
Example: GradientTolerance=eps
Data Types: single | double
Size of the history buffer for the Hessian approximation, specified as a positive
integer. At each iteration, the software constructs the Hessian using statistics from
the latest HessianHistorySize iterations.
Example: HessianHistorySize=10
Data Types: single | double
Maximal number of iterations in the training process for each quantile, specified as a positive integer.
Example: IterationLimit=1e7
Data Types: single | double
Other Regression Options
Categorical predictors list, specified as one of the values in this table. The descriptions assume that the predictor data has observations in rows and predictors in columns.
| Value | Description |
|---|---|
| Vector of positive integers |
Each entry in the vector is an index value indicating that the corresponding predictor is
categorical. The index values are between 1 and If |
| Logical vector |
A |
| Character matrix | Each row of the matrix is the name of a predictor variable. The names must match the entries in PredictorNames. Pad the names with extra blanks so each row of the character matrix has the same length. |
| String array or cell array of character vectors | Each element in the array is the name of a predictor variable. The names must match the entries in PredictorNames. |
"all" | All predictors are categorical. |
By default, if the
predictor data is in a table (Tbl), fitrqlinear
assumes that a variable is categorical if it is a logical vector, categorical vector, character
array, string array, or cell array of character vectors. If the predictor data is a matrix
(X), fitrqlinear assumes that all predictors are
continuous. To identify any other predictors as categorical predictors, specify them by using
the CategoricalPredictors name-value argument.
For the identified categorical predictors, fitrqlinear creates
dummy variables using two different schemes, depending on whether a categorical variable
is unordered or ordered. For an unordered categorical variable,
fitrqlinear creates one dummy variable for each level of the
categorical variable. For an ordered categorical variable,
fitrqlinear creates one less dummy variable than the number of
categories. For details, see Automatic Creation of Dummy Variables.
Example: CategoricalPredictors="all"
Data Types: single | double | logical | char | string | cell
Predictor variable names, specified as a string array of unique names or cell array of unique
character vectors. The functionality of PredictorNames depends on the
way you supply the training data.
If you supply
XandY, then you can usePredictorNamesto assign names to the predictor variables inX.The order of the names in
PredictorNamesmust correspond to the predictor order inX. Assuming thatXhas the default orientation, with observations in rows and predictors in columns,PredictorNames{1}is the name ofX(:,1),PredictorNames{2}is the name ofX(:,2), and so on. Also,size(X,2)andnumel(PredictorNames)must be equal.By default,
PredictorNamesis{'x1','x2',...}.
If you supply
Tbl, then you can usePredictorNamesto choose which predictor variables to use in training. That is,fitrqlinearuses only the predictor variables inPredictorNamesand the response variable during training.PredictorNamesmust be a subset ofTbl.Properties.VariableNamesand cannot include the name of the response variable.By default,
PredictorNamescontains the names of all predictor variables.A good practice is to specify the predictors for training using either
PredictorNamesorformula, but not both.
Example: PredictorNames=["SepalLength","SepalWidth","PetalLength","PetalWidth"]
Data Types: string | cell
Response variable name, specified as a character vector or string scalar.
If you supply
Y, then you can useResponseNameto specify a name for the response variable.If you supply
ResponseVarNameorformula, then you cannot useResponseName.
Example: ResponseName="response"
Data Types: char | string
Function for transforming raw response values, specified as a function handle or
function name. The default is "none", which means
@(y)y, or no transformation. The function should accept a vector
(the original response values) and return a vector of the same size (the transformed
response values).
Example: Suppose you create a function handle that applies an exponential
transformation to an input vector by using myfunction = @(y)exp(y).
Then, you can specify the response transformation as
ResponseTransform=myfunction.
Data Types: char | string | function_handle
Since R2025a
Option to perform computations in parallel using a parallel pool of workers, specified as one of these values:
false(0) — Run in serial on the MATLAB client.true(1) — Use a parallel pool if one is open or if MATLAB can automatically create one. If a parallel pool is not available, run in serial on the MATLAB client.
If you do not have a parallel pool open and automatic pool creation is enabled, MATLAB opens a pool using the default cluster profile. To use a parallel pool to run computations in MATLAB, you must have Parallel Computing Toolbox™. For more information, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
Tip
Set UseParallel to true when the
Quantiles value contains multiple quantiles.
fitrqlinear performs all computations in serial when there is
only one quantile.
Example: UseParallel=true
Data Types: single | double | logical | char | string
Observation weights, specified as a nonnegative numeric vector or the name of a variable in Tbl. The software weights each observation in X or Tbl with the corresponding value in Weights. The length of Weights must equal the number of observations in X or Tbl.
If you specify the input data as a table Tbl, then Weights can be the name of a variable in Tbl that contains a numeric vector. In this case, you must specify Weights as a character vector or string scalar. For example, if the weights vector W is stored as Tbl.W, then specify it as "W". Otherwise, the software treats all columns of Tbl, including W, as predictors when training the model.
By default, Weights is ones(n,1), where n is the number of observations in X or Tbl.
fitrqlinear normalizes the weights to sum to 1.
Data Types: single | double | char | string
Cross-Validation Options
Since R2025a
Flag to train a cross-validated model, specified as "on" or
"off".
If you specify "on", then the software trains a cross-validated
model with 10 folds.
You can override this cross-validation setting using the
CVPartition, Holdout,
KFold, or Leaveout name-value argument.
You can use only one cross-validation name-value argument at a time to create a
cross-validated model.
Alternatively, cross-validate later by passing Mdl to the
crossval function.
Example: CrossVal="on"
Data Types: char | string
Since R2025a
Cross-validation partition, specified as a cvpartition object that specifies the type of cross-validation and the
indexing for the training and validation sets.
To create a cross-validated model, you can specify only one of these four
name-value arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: Suppose you create a random partition for 5-fold cross-validation on 500
observations by using cvp = cvpartition(500,KFold=5). Then, you can
specify the cross-validation partition by setting
CVPartition=cvp.
Since R2025a
Fraction of the data used for holdout validation, specified as a scalar value in
the range (0,1). If you specify Holdout=p, then the software
completes these steps:
Randomly select and reserve
p*100% of the data as validation data, and train the model using the rest of the data.Store the compact trained model in the
Trainedproperty of the cross-validated model.
To create a cross-validated model, you can specify only one of these four
name-value arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: Holdout=0.1
Data Types: double | single
Since R2025a
Number of folds to use in the cross-validated model, specified as a positive
integer value greater than 1. If you specify KFold=k, then the
software completes these steps:
Randomly partition the data into
ksets.For each set, reserve the set as validation data, and train the model using the other
k– 1 sets.Store the
kcompact trained models in ak-by-1 cell vector in theTrainedproperty of the cross-validated model.
To create a cross-validated model, you can specify only one of these four
name-value arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: KFold=5
Data Types: single | double
Since R2025a
Leave-one-out cross-validation flag, specified as "on" or
"off". If you specify Leaveout="on", then for
each of the n observations (where n is the
number of observations, excluding missing observations, specified in the
NumObservations property of the model), the software completes
these steps:
Reserve the one observation as validation data, and train the model using the other n – 1 observations.
Store the n compact trained models in an n-by-1 cell vector in the
Trainedproperty of the cross-validated model.
To create a cross-validated model, you can specify only one of these four
name-value arguments: CVPartition, Holdout,
KFold, or Leaveout.
Example: Leaveout="on"
Data Types: char | string
Note
You cannot use any cross-validation name-value argument together with the
OptimizeHyperparameters name-value argument. You can modify the
cross-validation for OptimizeHyperparameters only by using the
HyperparameterOptimizationOptions name-value argument.
Hyperparameter Optimization Options
Since R2025a
Parameters to optimize, specified as one of the following:
"none"— Do not optimize."auto"— Use["Lambda","Standardize"]."all"— Optimize all eligible parameters.String array or cell array of eligible parameter names.
Vector of
optimizableVariableobjects, typically the output ofhyperparameters.
You can optimize hyperparameters only when creating a quantile regression model
with one quantile (that is, the Quantiles name-value argument has
one element).
The optimization attempts to minimize the cross-validation loss
(error) for fitrqlinear by varying the parameters. To control the
cross-validation type and other aspects of the optimization, use the
HyperparameterOptimizationOptions name-value argument. When you use
HyperparameterOptimizationOptions, you can use the (compact) model size
instead of the cross-validation loss as the optimization objective by setting the
ConstraintType and ConstraintBounds options.
Note
The values of OptimizeHyperparameters override any values you
specify using other name-value arguments. For example, setting
OptimizeHyperparameters to "auto" causes
fitrqlinear to optimize hyperparameters corresponding to the
"auto" option and to ignore any specified values for the
hyperparameters.
The eligible parameters for fitrqlinear are:
Lambda—fitrqlinearoptimizesLambdaover log-scaled values in the range[1e-5/NumObservations,1e5/NumObservations].Standardize—fitrqlinearoptimizesStandardizeover the two values[true,false].
Set nondefault parameters by passing a vector of
optimizableVariable objects that have nondefault values. For
example:
load carsmall params = hyperparameters("fitrqlinear",[Horsepower,Weight],MPG); params(1).Range = [1e-3,2e4];
Pass params as the value of
OptimizeHyperparameters.
By default, the iterative display appears at the command line,
and plots appear according to the number of hyperparameters in the optimization. For the
optimization and plots, the objective function is log(1 + cross-validation loss). To control the iterative display, set the Verbose option
of the HyperparameterOptimizationOptions name-value argument. To control
the plots, set the ShowPlots option of the
HyperparameterOptimizationOptions name-value argument.
Example: OptimizeHyperparameters="auto"
Since R2025a
Options for optimization, specified as a HyperparameterOptimizationOptions object or a structure. This argument
modifies the effect of the OptimizeHyperparameters name-value
argument. If you specify HyperparameterOptimizationOptions, you
must also specify OptimizeHyperparameters. All the options listed
in the following table are optional. However, you must set
ConstraintBounds and ConstraintType to return
AggregateOptimizationResults. The options that you can set in a
structure are the same as those in the
HyperparameterOptimizationOptions object.
| Option | Values | Default |
|---|---|---|
Optimizer |
| "bayesopt" |
ConstraintBounds | Constraint bounds for N optimization problems,
specified as an N-by-2 numeric matrix or
| [] |
ConstraintTarget | Constraint target for the optimization problems, specified as
| If you specify ConstraintBounds and
ConstraintType, then the default value is
"matlab". Otherwise, the default value is
[]. |
ConstraintType | Constraint type for the optimization problems, specified as
| [] |
AcquisitionFunctionName | Type of acquisition function:
Acquisition functions whose names include
| "expected-improvement-per-second-plus" |
MaxObjectiveEvaluations | Maximum number of objective function evaluations. If you specify multiple
optimization problems using ConstraintBounds, the value of
MaxObjectiveEvaluations applies to each optimization
problem individually. | 30 for "bayesopt" and
"randomsearch", and the entire grid for
"gridsearch" |
MaxTime | Time limit for the optimization, specified as a nonnegative real
scalar. The time limit is in seconds, as measured by | Inf |
NumGridDivisions | For Optimizer="gridsearch", the number of values in each
dimension. The value can be a vector of positive integers giving the number of
values for each dimension, or a scalar that applies to all dimensions. The
software ignores this option for categorical variables. | 10 |
ShowPlots | Logical value indicating whether to show plots of the optimization progress.
If this option is true, the software plots the best observed
objective function value against the iteration number. If you use Bayesian
optimization (Optimizer="bayesopt"), the
software also plots the best estimated objective function value. The best
observed objective function values and best estimated objective function values
correspond to the values in the BestSoFar (observed) and
BestSoFar (estim.) columns of the iterative display,
respectively. You can find these values in the properties ObjectiveMinimumTrace and EstimatedObjectiveMinimumTrace of
Mdl.HyperparameterOptimizationResults. If the problem
includes one or two optimization parameters for Bayesian optimization, then
ShowPlots also plots a model of the objective function
against the parameters. | true |
SaveIntermediateResults | Logical value indicating whether to save the optimization results. If this
option is true, the software overwrites a workspace variable
named "BayesoptResults" at each iteration. The variable is a
BayesianOptimization object. If you
specify multiple optimization problems using
ConstraintBounds, the workspace variable is an AggregateBayesianOptimization object named
"AggregateBayesoptResults". | false |
Verbose | Display level at the command line:
For details, see the | 1 |
UseParallel | Logical value indicating whether to run the Bayesian optimization in parallel, which requires Parallel Computing Toolbox. Due to the nonreproducibility of parallel timing, parallel Bayesian optimization does not necessarily yield reproducible results. For details, see Parallel Bayesian Optimization. | false |
Repartition | Logical value indicating whether to repartition the cross-validation at
every iteration. If this option is A value of
| false |
| Specify only one of the following three options. | ||
CVPartition | cvpartition object created by cvpartition | KFold=5 if you do not specify a
cross-validation option |
Holdout | Scalar in the range (0,1) representing the holdout
fraction | |
KFold | Integer greater than 1 | |
Example: HyperparameterOptimizationOptions=struct(UseParallel=true)
Output Arguments
Trained quantile linear regression model, returned as a RegressionQuantileLinear object, a RegressionPartitionedQuantileModel object, or a cell array of model objects.
If you set any of the name-value arguments
CrossVal,CVPartition,Holdout,KFold, orLeaveout, thenMdlis aRegressionPartitionedQuantileModelobject.If you specify
OptimizeHyperparametersand set theConstraintTypeandConstraintBoundsoptions ofHyperparameterOptimizationOptions, thenMdlis an N-by-1 cell array of model objects, where N is equal to the number of rows inConstraintBounds. If none of the optimization problems yields a feasible model, then each cell array value is[].Otherwise,
Mdlis aRegressionQuantileLinearmodel object.
To reference properties of a model object, use dot notation.
Since R2025a
Aggregate optimization results for multiple optimization problems, returned as an
AggregateBayesianOptimization object. To return
AggregateOptimizationResults, you must specify
OptimizeHyperparameters and
HyperparameterOptimizationOptions. You must also specify the
ConstraintType and ConstraintBounds options
of HyperparameterOptimizationOptions. For an example that shows how
to produce this output, see Hyperparameter Optimization with Multiple Constraint Bounds.
Tips
You can use the α/2 and 1 – α/2 quantiles to create a prediction interval that captures an estimated 100*(1 – α) percent of the variation in the response. For an example, see Create Prediction Interval Using Quantiles.
You can use quantile regression models to fit models that are robust to outliers. For an example, see Fit Regression Models to Data with Outliers.
Extended Capabilities
To run in parallel, set UseParallel in one of these ways:
Fit regression models for the quantiles in parallel by setting the
UseParallelname-value argument totrue.Perform this type of parallelization when you have multiple quantiles (that is, when
Quantileshas more than one element).Optimize in parallel by using the
UseParallel=trueoption in theHyperparameterOptimizationOptionsname-value argument.Perform this type of parallelization when you have one quantile (that is, when
Quantileshas one element). For more information on parallel hyperparameter optimization, see Parallel Bayesian Optimization.
For general information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
Version History
Introduced in R2024bWhen you use fitrqlinear, you can optimize a model, cross-validate
a model, or train a model in parallel.
To optimize the hyperparameters of a quantile regression model, specify the
OptimizeHyperparametersname-value argument.To cross-validate a quantile regression model, specify one of these name-value arguments:
CrossVal,CVPartition,Holdout,KFold, orLeaveout. Alternatively, create a full quantile regression model and then use thecrossvalobject function.To train a quantile linear regression model in parallel, specify the
UseParallelname-value argument astrue. To use a parallel pool to run computations in MATLAB, you must have Parallel Computing Toolbox.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Sélectionner un site web
Choisissez un site web pour accéder au contenu traduit dans votre langue (lorsqu'il est disponible) et voir les événements et les offres locales. D’après votre position, nous vous recommandons de sélectionner la région suivante : .
Vous pouvez également sélectionner un site web dans la liste suivante :
Comment optimiser les performances du site
Pour optimiser les performances du site, sélectionnez la région Chine (en chinois ou en anglais). Les sites de MathWorks pour les autres pays ne sont pas optimisés pour les visites provenant de votre région.
Amériques
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)