incrementalRobustRandomCutForest
Description
The incrementalRobustRandomCutForest function creates an
incrementalRobustRandomCutForest model object, which represents a robust random cut forest
(RRCF) model for incremental anomaly detection.
Unlike other Statistics and Machine Learning Toolbox™ model objects, incrementalRobustRandomCutForest can be called directly. Also,
you can specify learning options, such as the number of robust random cut trees, the
contamination fraction in the training data, and whether to standardize the predictor data
before fitting the model to data. After you create an incrementalRobustRandomCutForest object, it
is prepared for incremental learning (see Incremental Learning for Anomaly Detection).
incrementalRobustRandomCutForest is best suited for incremental learning. For a traditional
approach to anomaly detection when all the data is provided in advance, see rrcforest.
Creation
You can create an incrementalRobustRandomCutForest model object in several ways:
Call the function directly — Configure incremental learning options, or specify learner-specific options, by calling
incrementalRobustRandomCutForestdirectly. This approach is best when you do not have data yet or you want to start incremental learning immediately.Convert a traditionally trained model — To initialize a RRCF model for incremental learning using the model parameters and hyperparameters of a trained model object, you can convert the traditionally trained model to an
incrementalRobustRandomCutForestmodel object by passing it to theincrementalLearnerfunction.Call an incremental learning function —
fitaccepts a configuredincrementalRobustRandomCutForestmodel object and data as input, and returns anincrementalRobustRandomCutForestmodel object updated with information learned from the input model and data.
Syntax
Description
forest = incrementalRobustRandomCutForestforest for anomaly detection
with default parameters. Properties of a default model contain placeholders for unknown
model parameters. You must train a default model before you can use it to detect
anomalies.
forest = incrementalRobustRandomCutForest(Name,Value)incrementalRobustRandomCutForest(ContaminationFraction=0.1,ScoreWarmupPeriod=1000)
sets the anomaly contamination fraction to 0.1 and the score warm-up
period to 1000.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: incrementalRobustRandomCutForest(StandardizeData=true)
specifies to standardize the predictor data.
Flag to standardize the predictor data, specified as a numeric or logical 1
(true) or 0 (false).
If you set StandardizeData=true, the incrementalRobustRandomCutForest function centers and scales each predictor variable (X or Tbl) by the corresponding column mean and standard deviation. The function does not standardize the data contained in the dummy variable columns generated for categorical predictors.
Example: StandardizeData=true
Data Types: logical
Options for computing in parallel and setting random streams, specified as a
structure. Create the Options structure using statset. This table lists the option fields and their
values.
| Field Name | Value | Default |
|---|---|---|
UseParallel | Set this value to true to run computations in
parallel. | false |
UseSubstreams | Set this value to To compute
reproducibly, set | false |
Streams | Specify this value as a RandStream object or
cell array of such objects. Use a single object except when the
UseParallel value is true
and the UseSubstreams value is
false. In that case, use a cell array that
has the same size as the parallel pool. | If you do not specify Streams, then
incrementalRobustRandomCutForest uses the default stream or
streams. |
Note
You need Parallel Computing Toolbox™ to run computations in parallel.
Example: Options=statset(UseParallel=true,UseSubstreams=true,Streams=RandStream("mlfg6331_64"))
Data Types: struct
Warm-up period before score computation and anomaly detection, specified as a
nonnegative integer. This option specifies the number of observations used by the
incremental fit function to train the model and estimate the
score threshold.
Note
When processing observations during the score warm-up period, the software ignores observations that contain missing values for all predictors.
Example:
ScoreWarmupPeriod=200
Data Types: single | double
Running window size used to estimate the score threshold
(ScoreThreshold), specified as a positive integer. The
default ScoreWindowSize value is
1000.
If ScoreWindowSize is greater than the number of observations
in the training data, the software determines ScoreThreshold by
subsampling from the training data. Otherwise, ScoreThreshold is
set to forest.ScoreThreshold.
Example:
ScoreWindowSize=100
Data Types: single | double
Properties
You can set most properties by using name-value argument syntax when you call
incrementalRobustRandomCutForest directly. You can set some properties when you call
incrementalLearner to convert a traditionally trained model object. You
cannot set the properties Mu,
NumTrainingObservations, ScoreThreshold,
Sigma, and IsWarm.
This property is read-only.
List of categorical predictors, specified as one of the values in this table.
| Value | Description |
|---|---|
| Vector of positive integers | Each entry in the vector is an index value indicating that the corresponding predictor is categorical. The index values are between 1 and If |
| Logical vector | A |
| Character matrix | Each row of the matrix is the name of a predictor variable. The names must match the entries
in PredictorNames. Pad the names
with extra blanks so each row of the character matrix has the same
length. |
| String array or cell array of character vectors | Each element in the array is the name of a predictor variable. The names must match the entries in PredictorNames. |
"all" | All predictors are categorical. |
Data Types: single | double | logical | char | string | cell
This property is read-only.
Collusive displacement calculation method, specified as "maximal"
or "average".
The incrementalRobustRandomCutForest function finds the maximum change
("maximal") or the average change ("average") in
model complexity for each tree, and computes the collusive displacement (anomaly score)
for each observation.
Data Types: char | string
This property is read-only.
Fraction of anomalies in the training data, specified as a numeric scalar in the
range [0,1].
If the
ContaminationFractionvalue is0, thenincrementalRobustRandomCutForesttreats all training observations as normal observations, and sets theScoreThresholdvalue to the maximum anomaly score value of the training data.If the
ContaminationFractionvalue is in the range (0,1], thenincrementalRobustRandomCutForestdetermines theScoreThresholdvalue so that the function detects the specified fraction of training observations as anomalies.
The default ContaminationFraction value depends on how you
create the model:
If you convert a traditionally trained model to create
forest, thenContaminationFractionis specified by the corresponding property of the traditionally trained model.If you create
forestby callingincrementalRobustRandomCutForestdirectly, then you can specifyContaminationFractionby using name-value argument syntax. If you do not specify the value, then the default value is0.
Data Types: single | double
This property is read-only.
Number of observations processed by the incremental learner to estimate hyperparameters before training, specified as a nonnegative integer.
When processing observations during the estimation period, the software ignores observations that have missing values for all predictors.
If you specify a positive
EstimationPeriodandStandardizeDataisfalse,incrementalRobustRandomCutForestforcesEstimationPeriodto0.If
forestis prepared for incremental learning (all hyperparameters required for training are specified),incrementalRobustRandomCutForestforcesEstimationPeriodto0.If
forestis not prepared for incremental learning andStandardizeDataistrue,incrementalRobustRandomCutForestsetsEstimationPeriodto1000and estimates the unknown hyperparameters.
For more details, see Estimation Period.
Data Types: single | double
This property is read-only.
Flag indicating whether the incremental fitting function fit returns
scores and detects anomalies after training the model, specified as a numeric or logical
0 (false) or 1
(true).
The incremental model forest is warm
(IsWarm becomes true) after the
fit function fits the incremental model to
ScoreWarmupPeriod observations.
You cannot specify IsWarm directly.
Data Types: logical
This property is read-only.
Predictor means of the training data, specified as a numeric vector.
If you specify
StandardizeData=true:The length of
Muis equal to the number of predictors.If you set
StandardizeData=false, thenMuis an empty vector ([]).
You cannot specify Mu directly.
Data Types: single | double
This property is read-only.
Number of robust random cut trees (trees in the RRCF model), specified as a positive integer scalar.
Data Types: single | double
This property is read-only.
Number of observations to draw from the training data without replacement for each robust random cut tree (tree in the RRCF model), specified as a positive integer scalar greater than or equal to 3.
Data Types: single | double
This property is read-only.
Size of historical data that pertains to the RRCF model's knowledge, specified as a positive integer scalar.
Data Types: single | double
This property is read-only.
Number of predictor variables, specified as a nonnegative numeric scalar.
The default NumPredictors value depends on how you create the model:
If you convert a traditionally trained model to create
forest,NumPredictorsis specified by the corresponding property of the traditionally trained model.If you create
forestby callingincrementalRobustRandomCutForestdirectly, you can specifyNumPredictorsby using name-value argument syntax. If you do not specify the value, then the default value is0, and incremental fitting functions inferNumPredictorsfrom the predictor data during training.
Data Types: double
This property is read-only.
Number of observations fit to the incremental model forest,
specified as a nonnegative numeric scalar. NumTrainingObservations
increases when you pass forest and training data to
fit outside of the estimation period.
When fitting the model, the software ignores observations that have missing values for all predictors.
If you convert a traditionally trained model to create
forest,incrementalRobustRandomCutForestdoes not add the number of observations fit to the traditionally trained model toNumTrainingObservations.
You cannot specify NumTrainingObservations directly.
Data Types: double
Observation removal method, specified as "oldest",
"timedecaying", or "random". When the robust
random cut trees reach their capacity, the software removes old observations to
accommodate the most recent data.
| Value | Description |
|---|---|
| Oldest observations are removed first. |
| Observations are removed randomly in a weighted fashion. Older observations have a higher probability of being removed first. |
| Observations are removed in random order. |
Data Types: string | char
This property is read-only.
Predictor variable names, specified as a string array of unique names or cell array of
unique character vectors. The functionality of PredictorNames depends
on how you supply the predictor data.
If you supply
Tbl, then you can usePredictorNamesto specify which predictor variables to use. That is,incrementalRobustRandomCutForestuses only the predictor variables inPredictorNames.PredictorNamesmust be a subset ofTbl.Properties.VariableNames.By default,
PredictorNamescontains the names of all predictor variables inTbl.
If you supply
X, then you can usePredictorNamesto assign names to the predictor variables inX.The order of the names in
PredictorNamesmust correspond to the column order ofX. That is,PredictorNames{1}is the name ofX(:,1),PredictorNames{2}is the name ofX(:,2), and so on. Also,size(X,2)andnumel(PredictorNames)must be equal.By default,
PredictorNamesis{"x1","x2",...}.
Data Types: string | cell
This property is read-only.
Threshold for the anomaly score used to detect anomalies, specified as a nonnegative
integer. incrementalRobustRandomCutForest detects observations with scores above the
threshold as anomalies.
The default ScoreThreshold value depends on how you create the model:
If you convert a traditionally trained model object to create
forest, thenScoreThresholdis specified by the corresponding property value of the object.Otherwise, the default value is
0.
ScoreThreshold has the value 0 until the number of observations
reaches the ScoreWarmupPeriod value. After that, the software
updates the ScoreThreshold with every new observation.
You cannot specify ScoreThreshold directly.
Data Types: single | double
This property is read-only.
Warm-up period before score computation and anomaly detection, specified as a
nonnegative integer. This value is the number of observations used by the incremental
fit function to train the model and estimate the score
threshold.
When processing observations during the score warm-up period, the software ignores observations that have missing values for all predictors.
You can return scores and detect anomalies during the warm-up period by calling
isanomalydirectly.
The default ScoreWarmupPeriod value depends on how you create
the model:
If you convert a traditionally trained model to create
forest, theScoreWarmupPeriodname-value argument of theincrementalLearnerfunction sets this property.Otherwise, the default value is
0.
Data Types: single | double
This property is read-only.
Running window size for ScoreThreshold estimation, specified as
a nonnegative integer. The software estimates the ScoreThreshold
value over a running window with a window size of
ScoreWindowSize.
The default ScoreWindowSize value depends on how you create the model:
If you convert a traditionally trained model to create
forest, theScoreWindowSizename-value argument of theincrementalLearnerfunction sets this property.Otherwise, the default value is
1000.
Data Types: double
This property is read-only.
Predictor standard deviations of the training data, specified as a numeric vector.
If you specify
StandardizeData=truewhen you train an incremental RRCF model usingfit:The
fitfunction does not standardize columns that contain categorical variables. The elements inSigmafor categorical variables containNaNvalues.The
isanomalyfunction standardizes the input data by using the predictor means inMuand standard deviations inSigma.
The length of
Sigmais equal to the number of predictors.If you set
StandardizeData=false, thenSigmais an empty vector ([]).
You cannot specify Sigma directly.
Object Functions
Examples
Create a default robust random cut forest model for incremental anomaly detection.
forest = incrementalRobustRandomCutForest; details(forest)
incrementalRobustRandomCutForest with properties:
CollusiveDisplacement: 'maximal'
NumLearners: 100
NumObservationsPerLearner: 256
ObservationRemoval: 'oldest'
NumObservationsToKeep: 256
Mu: []
Sigma: []
CategoricalPredictors: []
EstimationPeriod: 0
IsWarm: 0
ContaminationFraction: 0
NumTrainingObservations: 0
NumPredictors: 0
ScoreThreshold: 0
ScoreWarmupPeriod: 0
PredictorNames: {}
ScoreWindowSize: 1000
Methods, Superclasses
forest is an incrementalRobustRandomCutForest model object. All its properties are read-only. By default, the software sets the anomaly contamination fraction to 0 and the score warm-up period to 0. forest must be fit to data before you can use it to perform any other operations.
Load Data
Load the human activity data set and keep only the first 3000 observations. For details on the data set, enter Description at the command line.
load humanactivity.mat
feat = feat(1:3000,:);Fit Incremental Model and Detect Anomalies
Fit the incremental model forest to the data by using the fit function. Because ScoreWarmupPeriod = 0, fit returns scores and detects anomalies immediately after fitting the model for the first time. To simulate a data stream, fit the model in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
medianscore, the median score value of the data chunk, to see how it evolves during incremental learning.Store
allscores, the score values for the fitted observations.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the data chunk.
n = numel(feat(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); medianscore = zeros(nchunk,1); threshold = zeros(nchunk,1); numAnom = zeros(nchunk,1); allscores = []; % Incremental fitting rng(0,"twister"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; forest = fit(forest,feat(idx,:)); [isanom,scores] = isanomaly(forest,feat(idx,:)); medianscore(j) = median(scores); allscores = [allscores scores']; numAnom(j) = sum(isanom); threshold(j) = forest.ScoreThreshold; end
forest is an incrementalRobustRandomCutForest model object trained on all the data in the stream. The fit function fits the model to the data chunk, and the isanomaly function returns the observation scores and the indices of observations in the data chunk with scores above the score threshold value.
Analyze Incremental Model During Training
Plot the anomaly score for every observation.
plot(allscores,".-") xlabel("Observation") ylabel("Score")

At each iteration, the software calculates a score value for each observation in the data chunk. A low score value indicates a normal observation, and a high score value indicates an anomaly.
To see how the score threshold and median score per data chunk evolve during training, plot them on separate tiles.
figure tiledlayout(2,1); nexttile plot(medianscore,".-") ylabel("Median Score") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(threshold,".-") ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk])
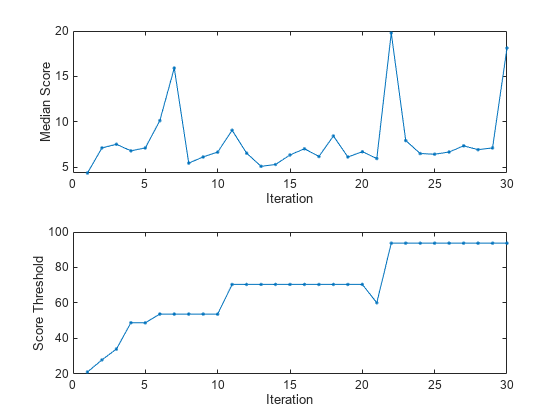
finalScoreThreshold=forest.ScoreThreshold
finalScoreThreshold = 93.7052
The median score fluctuates between 4 and 20. The anomaly score threshold has a value of 20 after the first iteration and steadily approaches a value of 94 by the 22nd iteration. Because ContaminationFraction = 0, incrementalRobustRandomCutForest treats all training observations as normal observations, and at each iteration sets the score threshold to the maximum score value in the data chunk.
totalAnomalies = sum(numAnom)
totalAnomalies = 0
No anomalies are detected at any iteration, because ContaminationFraction = 0.
Prepare an incremental robust random cut forest model by specifying an anomaly contamination fraction of 0.001, and standardize the data using an initial estimation period of 500 observations. Specify a score warm-up period of 1000 observations, during which the fit function updates the score threshold and trains the model but does not return scores or identify anomalies.
forest = incrementalRobustRandomCutForest(ContaminationFraction=0.001, ...
StandardizeData=true,ScoreWarmupPeriod=1000,EstimationPeriod=500);forest is an incrementalRobustRandomCutForest model object. All its properties are read-only. forest must be fit to data before you can use it to perform any other operations.
Load Data
Load the credit rating data stored in CreditRating_Historical.dat. Remove the ID column and the categorical variables.
creditrating = readtable("CreditRating_Historical.dat"); creditrating = removevars(creditrating,["ID","Industry","Rating"]);
The fit function of incrementalRobustRandomCutForest does not use observations with missing values. Remove missing values in the data sets to reduce memory consumption and speed up training.
creditrating = rmmissing(creditrating);
Fit Incremental Model and Detect Anomalies
Fit the incremental model to the data by using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. Because EstimationPeriod = 500 and ScoreWarmupPeriod = 1000, fit only returns scores and detects anomalies after 15 iterations. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
meanscore, the mean score value of the data chunk, to see how it evolves during incremental learning.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the chunk, to see how it evolves during incremental learning.
n = numel(creditrating(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); meanscore = zeros(nchunk,1); threshold = zeros(nchunk,1); numAnom = zeros(nchunk,1); % Incremental fitting rng(0,"twister"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [forest,tf,scores] = fit(forest,creditrating(idx,:)); meanscore(j) = mean(scores); numAnom(j) = sum(tf); threshold(j) = forest.ScoreThreshold; end
forest is an incrementalRobustRandomCutForest model object trained on all the data in the stream.
Analyze Incremental Model During Training
To see how the mean score, score threshold and number of detected anomalies per chunk evolve during training, plot them on separate tiles.
tiledlayout(3,1); nexttile plot(meanscore) ylabel("Mean Score") xlabel("Iteration") xlim([0 nchunk]) xline(forest.EstimationPeriod/numObsPerChunk,"r-.") xline((forest.EstimationPeriod+forest.ScoreWarmupPeriod)/numObsPerChunk,"r") nexttile plot(threshold) ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk]) xline(forest.EstimationPeriod/numObsPerChunk,"r-.") xline((forest.EstimationPeriod+forest.ScoreWarmupPeriod)/numObsPerChunk,"r") nexttile plot(numAnom,"+") ylabel("Anomalies") xlabel("Iteration") xlim([0 nchunk]) ylim([0 max(numAnom)+0.2]) xline(forest.EstimationPeriod/numObsPerChunk,"r-.") xline((forest.EstimationPeriod+forest.ScoreWarmupPeriod)/numObsPerChunk,"r")

During the estimation period, fit estimates means and standard deviations using the observations, and does not fit the model or update the score threshold. During the warm-up period, fit fits the model and updates the score threshold, but returns all scores as NaN and all anomaly values as false. After the warm-up period, fit returns the observation scores and the indices of observations with scores above the score threshold value. A small score value indicates a normal observation, and a large score value indicates an anomaly.
totalAnomalies=sum(numAnom)
totalAnomalies = 3
anomfrac= totalAnomalies/(n-forest.EstimationPeriod-forest.ScoreWarmupPeriod)
anomfrac = 0.0012
The software detects 3 anomalies after the warm-up and estimation periods. The contamination fraction after the estimation and warm-up periods is approximately 0.001.
More About
Incremental learning, or online learning, is a branch of machine learning concerned with processing incoming data from a data stream, possibly given little to no knowledge of the distribution of the predictor variables, aspects of the prediction or objective function (including tuning parameter values), or whether the observations contain anomalies. Incremental learning differs from traditional machine learning, where enough data is available to fit to a model, perform cross-validation to tune hyperparameters, and infer the predictor distribution.
Anomaly detection is used to identify unexpected events and departures from normal behavior. In situations where the full data set is not immediately available, or new data is arriving, you can use incremental learning for anomaly detection to incrementally train a model so it adjusts to the characteristics of the incoming data.
Given incoming observations, an incremental learning model for anomaly detection does the following:
Computes anomaly scores
Updates the anomaly score threshold
Detects data points above the score threshold as anomalies
Fits the model to the incoming observations
For more information, see Incremental Anomaly Detection with MATLAB.
Algorithms
During the estimation period, the incremental fitting function fit does not fit
the model. The function uses the first incoming EstimationPeriod observations
to estimate the predictor means (Mu) and standard deviations (Sigma). At the end of the
estimation period, the function updates the properties that store the
hyperparameters.
Estimation occurs only when:
EstimationPeriodis positive.forest.Muandforest.Sigmaare empty arrays[].Incremental fitting functions are configured to standardize predictor data (see Standardize Data).
Note
If you specify a positive EstimationPeriod and
StandardizeData is false, then
EstimationPeriod is reset to 0.
If incremental learning functions are configured to standardize predictor variables,
they do so using the means and standard deviations stored in the Mu and
Sigma properties of the incremental learning model
forest.
When you set
StandardizeData=trueand a positive estimation period (seeEstimationPeriod), andforest.Muandforest.Sigmaare empty, the incremental fit function estimates means and standard deviations using the estimation period observations.When the incremental fitting function estimates predictor means and standard deviations, the function computes weighted means and weighted standard deviations using the estimation period observations. Specifically, the function standardizes predictor j (xj) using
xj is predictor j, and xjk is observation k of predictor j in the estimation period.
wj is observation weight j.
The observation weights wj are all equal to one and cannot be specified.
References
[1] Guha, Sudipto, N. Mishra, G. Roy, and O. Schrijvers. "Robust Random Cut Forest Based Anomaly Detection on Streams," Proceedings of The 33rd International Conference on Machine Learning 48 (June 2016): 2712–21.
[2] Bartos, Matthew D., A. Mullapudi, and S. C. Troutman. "rrcf: Implementation of the Robust Random Cut Forest Algorithm for Anomaly Detection on Streams." Journal of Open Source Software 4, no. 35 (2019): 1336.
Extended Capabilities
To run in parallel, specify the Options name-value argument in the call to
this function and set the UseParallel field of the
options structure to true using
statset:
Options=statset(UseParallel=true)
For more information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
Version History
Introduced in R2023b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Sélectionner un site web
Choisissez un site web pour accéder au contenu traduit dans votre langue (lorsqu'il est disponible) et voir les événements et les offres locales. D’après votre position, nous vous recommandons de sélectionner la région suivante : .
Vous pouvez également sélectionner un site web dans la liste suivante :
Comment optimiser les performances du site
Pour optimiser les performances du site, sélectionnez la région Chine (en chinois ou en anglais). Les sites de MathWorks pour les autres pays ne sont pas optimisés pour les visites provenant de votre région.
Amériques
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)