Vous suivez désormais cette soumission
- Les mises à jour seront visibles dans votre flux de contenu suivi
- Selon vos préférences en matière de communication il est possible que vous receviez des e-mails
ecg-kit
Partager « ecg-kit »
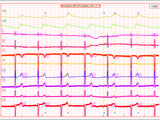
This toolbox is a collection of Matlab tools that I used, adapted or developed during my PhD and post-doc work with the Besicos group at University of Zaragoza, Spain and at the National Technological University of Buenos Aires, Argentina. The ecg-kit has tools for reading, processing and presenting results. The main feature of the this toolbox is the possibility to use several popular algorithms for ECG processing, such as:
- Algorithms from Physionet's WFDB software package
- QRS detectors, such as gqrs, wqrs, wavedet, ecgpuwave, Pan & Tompkins, EP limited
- Wavedet ECG delineator
- Pulse wave detectors as wabp and wavePPG
- a2hbc and EP limited heartbeat classifiers.
And other scritps for inspecting, correcting and reporting all these results.
Citation pour cette source
marianux (2026). ecg-kit (https://github.com/marianux/ecg-kit), GitHub. Extrait(e) le .
Compatibilité avec les versions de MATLAB
Plateformes compatibles
Windows macOS LinuxCatégories
- Industries > Medical Devices > Cardiology > ECG / EKG >
Découvrir Live Editor
Créez des scripts avec du code, des résultats et du texte formaté dans un même document exécutable.
common
- ADC2realunits.m
- ADC2units
- AUC_calc
- AnnotationFilterConvert
- Annotation_process
- BaselineWanderRemovalMedian
- BaselineWanderRemovalSplines
- CalcRRserieQuality
- CalcRRserieRatio
- CalculatePerformanceECGtaskQRSdet
- ConcatenateQRSdetectionPayloads
- Contents.m
- DelayedCovMat
- DisplayConfusionMatrix.m
- DisplayResults
- ECGformat
- ECGtask
- ECGtask
- ECGtask_Delineation_corrector
- ECGtask_ECG_delineation
- ECGtask_ECG_delineation_corrector
- ECGtask_PCA_proj_basis
- ECGtask_PPG_ABP_corrector
- ECGtask_PPG_ABP_detector
- ECGtask_QRS_corrector
- ECGtask_QRS_detection
- ECGtask_QRS_detections_post_process
- ECGtask_arbitrary_function
- ECGtask_do_nothing
- ECGtask_heartbeat_classifier
- ECGwrapper
- GTHTMLtable
- GetBestQRSdetections
- GetFunctionInvocation.m
- HasAdminPrivs
- MedianFilt
- MedianFiltSequence
- PeakDetection2
- PiCA
- PlotGlobalWaveMarks
- PlotWaveMarks
- PrctileFilt
- QRScorrector
- RR_calculation
- Seconds2HMS
- TaskPartition
- WFDB_command_prefix
- addpath_if_not_added
- adjust_string
- aip_detector
- aip_detector_concatenate
- alineate_positions
- arb_modmax
- arrow
- autovec_calculation
- autovec_calculation_robust
- bandpass_filter_design
- best_m_lead
- bxb
- calc_btime
- calc_co_ocurrences
- calc_correlation_gain
- calculateSeriesQuality
- cluster_data_with_EM_clust
- colvec
- combine_anns
- concat_QRS_detections
- cprintf
- deNaN_dataset
- default_concatenate_function
- default_finish_function
- default_start_function
- design_downsample_filter
- disp_option_enumeration
- disp_string_framed
- disp_string_title
- disp_xml_nodes
- ds2nfu
- ecgkit_wave_defs.m
- example_worst_ever_ECG_delineator
- example_worst_ever_QRS_detector
- exist_distributed_file
- getAnnNames
- get_ECG_idx_from_header
- get_PPG_ABP_idx_from_header
- get_QRS_from_payload
- get_segments_from_sequence
- get_time_from_xml_tag
- groupnlineup_waves
- init_WFDB_library
- init_ghostscript
- isAHAformat
- isHESformat
- isHL7aformat
- isISHNEformat
- isMatlab
- isOctave
- islater
- list_all_ECGtask
- list_recordings
- logit_function
- matformat_definitions.m
- matrix2positions
- max_index
- maximize
- merge_QRS_detections
- min_index
- mixartif
- modmax
- my_colormap
- myzerocros
- nanmeda
- pack_signal
- parse_pids
- plot_auc
- plot_ecg_heartbeat.m
- plot_ecg_mosaic
- plot_ecg_strip
- plot_roc
- positions2matrix
- ppval
- progress_bar
- progress_bar_ex.m
- qs_filter_design
- qs_wt.m
- rand_linespec
- read_310_format
- read_311_format
- read_AHA_ann
- read_AHA_format
- read_AHA_header
- read_ECG
- read_HES_ann
- read_HES_format
- read_HES_header
- read_Mortara
- read_Mortara_format
- read_Mortara_header
- read_hl7a_format
- read_ishne
- read_ishne_ann
- read_ishne_format
- read_ishne_header
- readheader
- reportECG
- resample_sequences
- rotateticklabel
- rowvec
- set_a_linespec
- set_rand_linespec
- similarity_calculation
- soft_range_conversion
- soft_set_difference
- soft_set_intersect
- soft_set_intersect
- soft_set_union
- sys_cmd_separation_string
- sys_command_strings
- tablas_y_constantes.m
- text_arrow
- text_line
- timeseries_explorer
- trim_ECG_header
- unique_w_tolerance
- wavedetMix
- woody_method
- writeannot.m
- writeheader
- xml_tag_value
common/LIBRA
- L1median
- adjustedboxplot
- adjustedoutlyingness
- adm
- agnes
- bagplot
- cda
- cdq
- chiqqplot
- clara
- classSVD
- clusplot
- cpca
- cpcr
- csimca
- csimpls
- cvMcd
- cvRobpca
- cvRpcr
- cvRsimpls
- daisy
- daplot
- ddplot
- diana
- distplot
- ellipsplot
- extractmcdregres
- fanny
- greatsort
- halfspacedepth
- hl
- kernelEVD
- libra_mahalanobis
- libra_ols
- lmc
- lsscatter
- ltsregres
- madc
- makeplot
- mc
- mcdcov
- mcdregres
- mcenter
- mlochuber
- mloclogist
- mlr
- mona
- mscalelogist
- normqqplot
- pam
- plotnumbers
- predict
- putlabel
- qn
- qnm
- randomset
- rapca
- rda
- regresdiagplot
- regresdiagplot3d
- removal
- removeObsMcd
- removeObsRobpca
- residualplot
- rmc
- robpca
- robpcaregres
- robstd
- rpcr
- rrmse
- rsimca
- rsimpls
- rsquared
- rstep
- scorediagplot
- screeplot
- simcaplot
- tree
- twopoints
- unimcd
- uniran
- updatecov
- weightmecov
common/a2hbc
common/a2hbc/scripts
- CollectResutls
- DisableControPanel.m
- DisplayConfiguration.m
- DoHouseKeeping
- ECGtask_classification_features_calc
- EnableControPanel.m
- ExpertUserInterface
- FindClusterExamples.m
- ParseUserControlPanelInput.m
- ResultsForAllDatabases
- UserControlPanel.m
- UserInteraction.m
- a2hbc_header.m
- a2hbc_main
- cluster_data_em
- position_waitbar.m
common/a2hbc/scripts/@featureMatrices
common/export_fig
- append_pdfs
- copyfig.m
- crop_borders
- eps2pdf.m
- export_fig.m
- fix_lines.m
- ghostscript.m
- im2gif
- isolate_axes.m
- pdf2eps.m
- pdftops
- print2array.m
- print2eps.m
- read_write_entire_textfile
- user_string
- using_hg2
common/kur
common/plot2svg
- demo_3d_plot2svg.m
- demo_svg_water
- plot2svg
- plot2svg
- simulink2svg
- svgAnimation
- svgBoundingBox
- svgClipping
- svgComposite
- svgDisplacementMap
- svgFlood
- svgGaussianBlur
- svgImage
- svgLuminanceToAlpha
- svgMorphology
- svgOffset
- svgSpecularLightingDistant
- svgTile
- svgTurbulence
- tutorial_filters
- tutorial_plot2svg
- tutorial_plot2svg_beta
common/ppg
common/prtools
- Contents.m
- Readme.m
- adaboostc
- affine
- arrfit
- averagec
- bagc.m
- bagcc.m
- baggingc
- band2obj.m
- bandsel.m
- bayesc.m
- bhatm.m
- bpxnc
- cat2data.m
- cdats.m
- checktoolbox.m
- chernoffm
- circles3d
- classc
- classd
- classim
- classnames
- cleval
- clevalb
- clevalf
- clevals
- closemess.m
- cmapm
- cnormc
- col2gray
- compute_kernel.m
- concatm.m
- confmat
- costm
- createdatafile.m
- crossval
- data2im
- datafiles.m
- dataim
- datasetconv.m
- datasetm.m
- datasets.m
- datfilt
- datgauss
- datunif
- dcsc
- define_mapping.m
- delfigs.m
- dipbin
- dipim
- dipimagecheck.m
- diplibwarn.m
- disnorm
- disperror
- distm
- distmaha
- doublem.m
- dps.m
- drbmc
- dtc
- dyadicm.m
- edicon.m
- emclust
- fdsc.m
- feat2lab.m
- feat2obj.m
- feateval
- featrank
- featsel.m
- featselb
- featself
- featseli
- featsellr
- featselm
- featselo
- featselp
- featselv.m
- featsetc.m
- featsetcc.m
- ffnc
- file2dset
- filtim
- filtm
- findclasses
- fisherc
- fisherm
- fixedcc
- font_size
- gauss
- gaussm
- gencirc
- genclass
- gendat
- gendatb
- gendatc
- gendatd
- gendatgauss
- gendath
- gendati.m
- gendatk
- gendatl
- gendatlin
- gendatm
- gendatp
- gendatr
- gendats
- gendats
- gendatsin
- gendatsinc
- gendatv
- gendatw.m
- genlab
- gensubsets.m
- getfeat
- getlab
- getname.m
- getopt_pars.m
- getwindows.m
- gpr
- gridsize
- gtm
- hclust
- histm.m
- im2feat
- im2obj
- im_bdilation
- im_berosion
- im_box
- im_bpropagation
- im_center
- im_dbr.m
- im_features
- im_fft
- im_fill_norm.m
- im_gauss
- im_gauss
- im_gaussf
- im_gray
- im_harris.m
- im_hist.m
- im_hist_equalize
- im_invert
- im_label
- im_maxf
- im_mean
- im_measure
- im_minf
- im_moments.m
- im_norm.m
- im_patch
- im_profile
- im_resize.m
- im_rotate
- im_scale
- im_select_blob
- im_skel
- im_skel_meas
- im_stat
- im_stretch
- im_threshold
- image_dbr
- immoments.m
- invsigm
- is_scalar.m
- iscomdset.m
- isdatafile
- isdataim
- isdataset
- isfeatim
- isgenerator
- ismapping
- isobjim
- isparallel
- issequential
- isstacked
- issym
- istrained
- isuntrained
- isvaldfile.m
- isvaldset.m
- kcentres
- kernelc.m
- kernelm
- klldc
- klm
- klms
- kmeans
- knn_map
- knnc
- knnm
- knnr
- ksmoothr
- labcmp.m
- labeld
- labelim
- lassor
- ldc
- libsvc.m
- libsvmcheck.m
- linearr
- lines5d
- linewidth
- lkc.m
- lmnc
- logdens.m
- loglc
- loso.m
- lssvc
- makeqld.m
- map
- mapex
- mapm
- mapping_task.m
- mappings.m
- mappingtools.m
- marksize
- matchcost
- matchlab
- matchlablist
- maxc
- mclassc
- mclassm.m
- mds
- mds_cs
- mds_init
- mds_stress
- mdsc.m
- meanc
- meancov
- medianc
- minc
- misval
- mlrc
- modeclust.m
- modeseek
- modselc
- mogc
- multi_labeling.m
- myfixedmapping
- naivebc.m
- naivebcc
- nbayesc
- neurc
- newfig
- newline
- newline
- nfminbnd
- nlabcmp
- nlabeld
- nlfisherm
- nmc
- nmsc
- nodatafile
- normal_map
- normm
- nu_svr.m
- nu_svro
- nulibsvc.m
- nusvc.m
- nusvo
- obj2feat.m
- out2.m
- outm.m
- parallel
- parsc
- parzen_map
- parzenc.m
- parzendc.m
- parzenm
- parzenml
- pca
- pcaklm
- pcam
- pcldc
- perc
- perlc
- pinvr.m
- pklibsvc.m
- plotc
- plotd
- plotdg
- plote
- plotf
- plotgtm.m
- plotm
- ploto
- plotr
- plotsom
- plottarget
- pls_apply
- pls_prepro
- pls_train
- pls_transform
- pls_updstruct
- plsm
- plsm
- polyc
- pr_newrb
- prarff.m
- prcov
- prcrossval
- prcursor
- prdata
- prdatafiles.m
- prdataset
- prdatasets.m
- prdownload.m
- preig
- prex_cleval.m
- prex_combining.m
- prex_confmat.m
- prex_cost.m
- prex_datafile.m
- prex_datasets.m
- prex_density.m
- prex_eigenfaces.m
- prex_logdens.m
- prex_matchlab.m
- prex_mcplot.m
- prex_mds.m
- prex_parzen.m
- prex_plotc.m
- prex_regr.m
- prex_silhouette_classification.m
- prex_soft.m
- prex_som.m
- prex_spatm.m
- prforum.m
- prglobal.m
- primport
- prinv
- prkmeans
- prload.m
- prmap
- prmemory
- prnews.m
- procm
- prodc
- proxm
- prpinv
- prplotsom.m
- prprogress.m
- prrank
- prroc
- prsvd
- prtest.m
- prtestc.m
- prtestf.m
- prtools4to5.m
- prtools_mod.m
- prtrace
- prtver.m
- prversion
- prwaitbar.m
- prwaitbarinit.m
- prwaitbarnext.m
- prwaitbaronce.m
- prwarning
- qdc
- quadrc
- randomforestc
- randreset
- rblibsvc.m
- rbnc
- rbsvc.m
- reducm
- regoptc.m
- reject
- rejectc.m
- rejectm
- remclass.m
- remoutl
- renumlab
- reorderclasses
- reorderdset
- resizem.m
- ridger
- rnnc
- roc
- rsquared
- rsscc.m
- runcells.m
- sammonm.m
- savedatafile.m
- scalem
- scatterd
- scatterdui
- scattern.m
- selclass
- seldat
- selectim.m
- sequential
- setbatch
- setdat
- setdata.m
- setdefaults.m
- setfeatsize
- setname.m
- shiftargin.m
- shiftop.m
- show.m
- showfigs.m
- sigm
- som
- spatm
- spirals
- stacked
- stamp_map.m
- statscheck.m
- statsdtc
- statsknnc
- statslinc
- statsnbc
- stumpc
- subsc
- svc
- svc_nu.m
- svcinfo.m
- svmr
- svo
- svo_nu
- testauc.m
- testc
- testcost
- testd
- testdatasize.m
- testk
- testn
- testp
- testr
- traincc
- trained_classifier.m
- trained_mapping.m
- tree_map
- treec
- tsnem
- txtread
- typp
- udc
- unitm
- unittm
- updstruct
- use_old_mapping_times.m
- userkernel.m
- vandermondem
- votec
- vpc
- weakc.m
- wvotec.m
common/prtools/@prdatafile
- abs.m
- addpostproc.m
- addpreproc.m
- and.m
- check12.m
- display.m
- dyadic.m
- end.m
- eq.m
- exp.m
- filenames.m
- find.m
- findfiles.m
- ge.m
- get.m
- getdata.m
- getdataset.m
- getfeatlab.m
- getfeatsize.m
- getfiles.m
- getpostproc.m
- getpreproc.m
- getsize.m
- gettype.m
- gt.m
- hist.m
- horzcat.m
- imread.m
- isempty.m
- isfinite.m
- isnan.m
- istype.m
- ldivide.m
- le.m
- log.m
- lt.m
- max.m
- mean.m
- median.m
- min.m
- minus.m
- mldivide.m
- mpower.m
- mrdivide.m
- mtimes.m
- ne.m
- not.m
- or.m
- plus.m
- power.m
- prdatafile.m
- prod.m
- rdivide.m
- readdatafile.m
- readdatafile2.m
- real.m
- set.m
- setdataset.m
- setfeatsize.m
- setfiles.m
- setpostproc.m
- setpreproc.m
- setraw.m
- settargets.m
- settype.m
- show.m
- sign.m
- size.m
- sqrt.m
- std.m
- struc.m
- subsasgn.m
- subsref.m
- sum.m
- times.m
- uminus.m
- uplus.m
- var.m
- vertcat.m
- xor.m
common/prtools/@prdataset
- abs.m
- addlabels.m
- addlablist.m
- addtargets.m
- and.m
- changelablist.m
- classsizes.m
- conj.m
- corrcoef.m
- crossval.m
- ctranspose.m
- cumsum.m
- curlablist.m
- dellablist.m
- det.m
- disp.m
- display.m
- double.m
- eig.m
- end.m
- eq.m
- exp.m
- find.m
- findfeatlab.m
- findident.m
- findlabels.m
- findnlab.m
- ge.m
- get.m
- getclassi.m
- getcost.m
- getdata.m
- getfeatdom.m
- getfeatlab.m
- getfeatsize.m
- getident.m
- getimheight.m
- getlabels.m
- getlablist.m
- getlablistnames.m
- getlabtype.m
- getname.m
- getnlab.m
- getobjsize.m
- getprior.m
- getsize.m
- gettargets.m
- getuser.m
- getversion.m
- gt.m
- hist.m
- horzcat.m
- hsv2rgb.m
- imsize.m
- inv.m
- invsig.m
- isempty.m
- isfinite.m
- islabtype.m
- isnan.m
- kmeans.m
- ldivide.m
- le.m
- length.m
- loadobj.m
- log.m
- lt.m
- max.m
- mean.m
- median.m
- min.m
- minus.m
- mldivide.m
- mpower.m
- mrdivide.m
- mtimes.m
- ndims.m
- ne.m
- not.m
- or.m
- pca.m
- pdist.m
- pinv.m
- plot.m
- plus.m
- power.m
- prdataset.m
- prforum_private.m
- prod.m
- rdivide.m
- real.m
- repmat.m
- rgb2hsv.m
- roc.m
- set.m
- setcost.m
- setdata.m
- setfeatdom.m
- setfeatlab.m
- setfeatsize.m
- setident.m
- setlabels.m
- setlablist.m
- setlablistnames.m
- setlabtype.m
- setname.m
- setnlab.m
- setobjsize.m
- setprior.m
- settargets.m
- setuser.m
- setversion.m
- show.m
- sign.m
- size.m
- sort.m
- sqrt.m
- std.m
- subsasgn.m
- subsref.m
- sum.m
- svd.m
- testfeatdom.m
- times.m
- uminus.m
- uplus.m
- var.m
- vertcat.m
- xor.m
common/prtools/@prmapping
- addpostproc.m
- and.m
- ctranspose.m
- disp.m
- display.m
- double.m
- end.m
- eq.m
- ge.m
- get.m
- getbatch.m
- getcost.m
- getdata.m
- getlabels.m
- getmapping_file.m
- getmapping_type.m
- getname.m
- getout_conv.m
- getscale.m
- getsize.m
- getsize_in.m
- getsize_out.m
- getuser.m
- gt.m
- horzcat.m
- isaffine.m
- isclassifier.m
- iscombiner.m
- isempty.m
- isfixed.m
- isfixed_cell.m
- isgenerator.m
- ldivide.m
- le.m
- lt.m
- minus.m
- mrdivide.m
- mtimes.m
- ne.m
- not.m
- or.m
- orth.m
- plot.m
- plotsom.m
- plus.m
- plus_org.m
- power.m
- prdataset.m
- prmapping.m
- rdivide.m
- set.m
- setcost.m
- setdata.m
- setlabels.m
- setmapping_file.m
- setmapping_type.m
- setname.m
- setout_conv.m
- setpostproc.m
- setscale.m
- setsize.m
- setsize_in.m
- setsize_out.m
- setuser.m
- show.m
- size.m
- subsasgn.m
- subsref.m
- times.m
- times_old.m
- uminus.m
- uplus.m
- vertcat.m
- xor.m
common/prtools/private
- bord
- covm
- expandd
- is_scalar.m
- isdipimage.m
- isint
- isint
- makeqld.m
- newline
- pd_check
- prdebug
- prmem
- resize
- strlab
common/prtools_addins
- CG_CLASSIFY
- CG_CLASSIFY_INIT
- classification_errors
- confmat_new
- countTransitions
- dbnFit
- dbnPredict
- dbnc
- distm_new
- distmaha_new
- emclust
- emclust_new
- feateval_new
- findoutl
- gaussm
- generalized_qdc
- getident_multids
- getnlab_multids
- hmmViterbiCM
- hmm_prtools
- labeld
- logistic.m
- loglc
- matchlab_new
- meancov_new
- minimize
- mogc
- my_remclass.m
- normal_map_new
- normal_map_new
- normalize
- nusvc.m
- nusvc_new.m
- nusvo
- nusvo_new
- plotc
- plotc
- plotm
- plotm
- prepareArgs
- process_options
- prroc_new
- prtools_mod.m
- prversion
- qdc_new
- qdc_new1
- rbmBB
- rbmPredict
- rbmVtoH
- rejectc_balanced.m
- rejectm_balanced
- roc
- roc_detection
- scatterd
- weighted_ldc
- weighted_qdc
common/prtools_addins/private
- bord
- covm
- expandd
- is_scalar.m
- isdipimage.m
- isint
- isint
- makeqld.m
- newline
- pd_check
- prdebug
- prmem
- resize
- strlab
common/wavedet
- SLR
- SLR2
- Tbeginauxiliar.m
- buscamin
- delineate3D
- delineateP3D
- fiducialf
- how_to_use.m
- isqrs
- isqrs
- locmax
- multiqrsend
- multiqrson
- nancov
- nanmax
- nanmean
- nanmedian
- nanmin
- nanstd
- nansum
- nanvar
- objfun2
- optimline
- picant
- picpost
- pos2ann
- posmat
- prepECG
- pwave_task1.m
- pwavef
- qrscandidatesnew
- qrswavef
- qspfilt5
- randsample
- readECG_v1
- reportECG
- rmanot.m
- searchbk
- searchoff
- searchon
- signaltest
- sincronizabeats
- t5.m
- twave3D
- twavef
- wavedet
- wavedet_3D
- wavedet_interface
- wavt
- wavt5
- zerocros
examples
Les versions qui utilisent la branche GitHub par défaut ne peuvent pas être téléchargées
| Version | Publié le | Notes de version | |
|---|---|---|---|
| 1.4.0.0 | Some minor fixes since v0.1.1 specially for MAC users.
|
|
|
| 1.2.0.0 | Nothing |
|
|
| 1.1.0.0 | More info in the toolbox web page ! http://marianux.github.io/ecg-kit/ or in the repository itself |
|
|
| 1.0.0.0 |
|

Sélectionner un site web
Choisissez un site web pour accéder au contenu traduit dans votre langue (lorsqu'il est disponible) et voir les événements et les offres locales. D’après votre position, nous vous recommandons de sélectionner la région suivante : .
Vous pouvez également sélectionner un site web dans la liste suivante :
Comment optimiser les performances du site
Pour optimiser les performances du site, sélectionnez la région Chine (en chinois ou en anglais). Les sites de MathWorks pour les autres pays ne sont pas optimisés pour les visites provenant de votre région.
Amériques
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)
Asie-Pacifique
- Australia (English)
- India (English)
- New Zealand (English)
- 中国
- 日本Japanese (日本語)
- 한국Korean (한국어)