fairnessThresholder
Description
fairnessThresholder searches for an optimal score threshold
to maximize accuracy while satisfying fairness bounds. For observations in the critical region
below the optimal threshold, the function adjusts the labels so that the fairness constraints
hold for the reference and nonreference groups in the sensitive attribute. After you create a
fairnessThresholder object, you can use the predict and
loss object
functions on new data to predict fairness labels and calculate the classification loss,
respectively.
Creation
Syntax
Description
fairnessMdl = fairnessThresholder(Mdl,Tbl,AttributeName,ResponseVarName)Mdl while
satisfying fairness bounds. The function tries a vector of thresholds for classifying
observations in the validation data table Tbl with the class labels
in the ResponseVarName table variable. For observations in the
critical region below the optimal threshold, the function adjusts the labels so that the
fairness constraints hold for the reference and nonreference groups in the
AttributeName sensitive attribute. For more information, see Reject Option-Based Classification.
fairnessMdl = fairnessThresholder(___,Name=Value)BiasMetric name-value argument.
Input Arguments
Binary classifier, specified as a full or compact classification model object or a function handle.
Full or compact model object — You can specify a full or compact classification model object, which has a
predictobject function. When you train a model, use a numeric matrix or table for the predictor data where rows correspond to individual observations.Supported Model Full or Compact Classification Model Object Discriminant analysis classifier ClassificationDiscriminant,CompactClassificationDiscriminantEnsemble of learners for classification ClassificationEnsemble,CompactClassificationEnsemble,ClassificationBaggedEnsembleGaussian kernel classification model using random feature expansion ClassificationKernelGeneralized additive model ClassificationGAM,CompactClassificationGAMk-nearest neighbor classifier ClassificationKNNLinear classification model ClassificationLinearNaive Bayes model ClassificationNaiveBayes,CompactClassificationNaiveBayesNeural network classifier ClassificationNeuralNetwork,CompactClassificationNeuralNetworkSupport vector machine classifier for binary classification ClassificationSVM,CompactClassificationSVMBinary decision tree for classification ClassificationTree,CompactClassificationTreeFunction handle — You can specify a function handle that accepts predictor data and returns a column vector containing a predicted score for each observation in the predictor data. Each predicted score must have a value between 0 and 1, where a score in the range [0, 0.5] corresponds to the negative class, and a score in the range (0.5, 1] corresponds to the positive class. You must specify the positive class using the
PositiveClassname-value argument.
Validation data set, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one variable. The table
must include all predictor variables used to train Mdl, the
sensitive attribute, and the response variable. The table can include additional
variables, such as observation weights. Multicolumn variables and cell arrays other
than cell arrays of character vectors are not allowed.
Data Types: table
Sensitive attribute name, specified as the name of a variable in
Tbl. You must specify AttributeName as a
character vector or a string scalar. For example, if the sensitive attribute is stored
as Tbl.Attribute, then specify it as
"Attribute".
The sensitive attribute must be a numeric vector, logical vector, character array, string array, cell array of character vectors, or categorical vector.
Data Types: char | string
Response variable name, specified as the name of a variable in
Tbl. You must specify ResponseVarName as a
character vector or a string scalar. For example, if the response variable is stored
as Tbl.Y, then specify it as "Y".
The response variable must be a numeric vector, logical vector, character array,
string array, cell array of character vectors, or categorical vector. The data type
must be the same as the data type of the response variable used to train
Mdl.
Data Types: char | string
Validation predictor data, specified as a numeric matrix. Each row of
X corresponds to one observation, and each column corresponds
to one predictor variable.
X,attribute, andYmust have the same number of rows.The columns of
Xmust have the same order as the predictor variables used to trainMdl.
Data Types: single | double
Sensitive attribute, specified as a numeric column vector, logical column vector, character array, string array, cell array of character vectors, or categorical column vector.
X,attribute, andYmust have the same number of rows.If
attributeis a character array, then each row of the array must correspond to a group in the sensitive attribute.
Data Types: single | double | logical | char | string | cell | categorical
Class labels, specified as a numeric column vector, logical column vector, character array, string array, cell array of character vectors, or categorical column vector.
X,attribute, andYmust have the same number of rows.If
Yis a character array, then each row of the array must correspond to a class label.The data type of
Ymust be the same as the data type of the response variable used to trainMdl.If
Mdlis a classification model object, then the distinct classes inYmust be a subset of the classes inMdl.ClassNames.
Data Types: single | double | logical | char | string | cell | categorical
Score threshold, specified as a numeric scalar.
fairnessThresholder adjusts the label for each observation
whose maximum score is less than the threshold value.
If
Mdlor itspredictobject function returns classification scores that are posterior probabilities, then specify athresholdvalue in the range [0.5, 1].If the
predictobject function ofMdlreturns classification scores in the range (–∞,∞), then specify a nonnegativethresholdvalue.
Data Types: single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: fairnessThresholder(Mdl,Tbl,"Gender","Smoker",BiasMetric="spd",BiasMetricRange=[–0.1
0.1]) specifies to find a score threshold so that the statistical parity
difference for the nonreference group in the Gender sensitive attribute
is in the range [–0.1, 0.1].
Bias metric to use as a fairness constraint during the threshold optimization, specified as one of the metric names in this table.
| Metric Name | Description |
|---|---|
"DisparateImpact" or "di"
(default) | Disparate impact (DI) |
"AverageAbsoluteOddsDifference" or
"aaod" | Average absolute odds difference (AAOD) |
"EqualOpportunityDifference" or
"eod" | Equal opportunity difference (EOD) |
"StatisticalParityDifference" or
"spd" | Statistical parity difference (SPD) |
For more information on the bias metric definitions, see Bias Metrics.
The fairnessThresholder function computes the bias metric
for the nonreference group (that is, the complement of
ReferenceGroups) and checks whether the value is within the
bias metric bounds (BiasMetricRange).
Example: BiasMetric="spd"
Example: BiasMetric="EqualOpportunityDifference"
Data Types: char | string
Bounds on the bias metric to use as constraints during the threshold optimization, specified as a two-element numeric vector. This table describes the supported bias metric values and the default bias metric bounds for each bias metric.
| Metric Name | Supported Bias Metric Values | Default BiasMetricRange Value |
|---|---|---|
"DisparateImpact" or
"di" | [0, ∞) | [0.8, 1.25] |
"AverageAbsoluteOddsDifference" or
"aaod" | [0, 1] | [0, 0.05] |
"EqualOpportunityDifference" or
"eod" | [–1, 1] | [–0.05, 0.05] |
"StatisticalParityDifference" or
"spd" | [–1, 1] | [–0.05, 0.05] |
The fairnessThresholder function computes the bias metric
(BiasMetric) for the nonreference group (that is, the
complement of ReferenceGroups) and checks whether the value is
within the bias metric bounds.
Example: BiasMetricRange=[-0.1 0.1]
Data Types: single | double
Groups in the sensitive attribute to use as the reference group when computing
bias metrics, specified as a scalar or a vector. By default,
fairnessThresholder chooses the most frequently occurring
group in the validation data as the reference group. Each element in the
ReferenceGroups value must have the same data type as the
sensitive attribute.
The function uses a technique designed for binary sensitive attributes that
contain a reference group and a nonreference group. Sensitive attribute groups not
in the ReferenceGroups value form the nonreference
group.
Example: ReferenceGroups=categorical(["Husband","Unmarried"])
Data Types: single | double | logical | char | string | cell | categorical
Label of the positive class, specified as a numeric scalar, logical scalar,
character vector, string scalar, cell array containing one character vector, or
categorical scalar. PositiveClass must have the same data type
as the true class label variable.
The default PositiveClass value is the second class of the
binary labels, according to the order returned by the unique function with the "sorted" option specified
for the true class label variable.
Example: PositiveClass=categorical(">50K")
Data Types: single | double | logical | char | string | cell | categorical
Loss to minimize during the threshold optimization, specified as
"classiferror", "classifcost", or a function
handle.
This table lists the available loss functions. Specify one using its corresponding character vector or string scalar.
| Value | Description | Equation |
|---|---|---|
"classifcost" | Observed misclassification cost |
|
"classiferror" | Misclassified rate in decimal |
|
C is the misclassification cost matrix, and I is the indicator function. If
Mdlis a classification model object, the misclassification cost matrix corresponds to theCostproperty ofMdl. IfMdlis a function handle, C is the default cost matrix, and the loss values for"classifcost"and"classiferror"are identical.yj is the true class label for observation j, and yj belongs to class kj.
is the class label with the maximal predicted score for observation j, and belongs to class .
n is the number of observations in the validation data set.
To specify a custom loss function, you must specify Mdl as
a classification model object. Use function handle notation
(@), where the function has
this form:lossfun
lossvalue = lossfun(Class,Score,Cost)
The output argument
lossvalueis a scalar.You specify the function name (
lossfun).Classis ann-by-Klogical matrix with rows indicating the class to which the corresponding observation belongs.nis the number of observations inTblorX, andKis the number of distinct classes in the response variable. The column order corresponds to the class order inMdl.ClassNames. CreateClassby settingClass(p,q) = 1, if observationpis in classq, for each row. Set all other elements of rowpto0.Scoreis ann-by-Knumeric matrix of classification scores. The column order corresponds to the class order inMdl.ClassNames.Scoreis a matrix of classification scores, similar to the output ofpredict.Costis aK-by-Knumeric matrix of misclassification costs. For example,Cost = ones(K) – eye(K)specifies a cost of0for correct classification and1for misclassification.
Example: LossFun="classifcost"
Data Types: char | string | function_handle
Maximum number of threshold values to evaluate during the threshold
optimization, specified as a positive integer.
fairnessThresholder uses a vector of
min(n,MaxNumThresholds) threshold values as part of the
optimization process, where n is the number of observations in
the validation data.
Example: MaxNumThresholds=250
Data Types: single | double
Properties
This property is read-only.
Binary classifier, returned as a full or compact classification model object or a function handle.
This property is read-only.
Sensitive attribute, returned as a variable name, numeric column vector, logical column vector, character array, cell array of character vectors, or categorical column vector.
If you use a table to create the
fairnessThresholderobject, thenSensitiveAttributeis the name of the sensitive attribute. The name is stored as a character vector.If you use a matrix to create the
fairnessThresholderobject, thenSensitiveAttributehas the same size and data type as the sensitive attribute used to create the object. (The software treats string arrays as cell arrays of character vectors.)
Data Types: single | double | logical | char | cell | categorical
This property is read-only.
Groups in the sensitive attribute to use as the reference group, returned as a scalar or vector. (The software treats string arrays as cell arrays of character vectors.)
The ReferenceGroups name-value argument sets this property.
Data Types: single | double | logical | char | cell | categorical
This property is read-only.
Name of the true class label variable, returned as a character vector containing the name of the response variable. (The software treats a string scalar as a character vector.)
If you specify the input argument
ResponseVarName, then its value determines this property.If you specify the input argument
Y, then the property value is'Y'.
Data Types: char
This property is read-only.
Label of the positive class, returned as a numeric scalar, logical scalar, character vector, cell array containing one character vector, or categorical scalar. (The software treats a string scalar as a character vector.)
The PositiveClass
name-value argument sets this property.
Data Types: single | double | logical | char | cell | categorical
This property is read-only.
Score threshold, returned as a numeric scalar. The score threshold is the optimal
score threshold derived by fairnessThresolder or the
threshold input argument value.
The ScoreThreshold property is empty when the original model
predictions already satisfy the fairness constraints or when all potential score
thresholds fail to satisfy the fairness constraints.
Data Types: single | double
This property is read-only.
Bias metric, returned as a character vector.
The BiasMetric
name-value argument sets this property.
Data Types: char
This property is read-only.
Bias metric value for the nonreference group, returned as a numeric scalar.
Sensitive attribute groups not in the ReferenceGroups value form
the nonreference group.
fairnessThresholder computes the bias metric value by using the
validation data set predictions, adjusted using the ScoreThreshold
value.
The BiasMetricValue property is empty when the original model
predictions already satisfy the fairness constraints or when all potential score
thresholds fail to satisfy the fairness constraints.
Data Types: double
This property is read-only.
Bounds on the bias metric, returned as a two-element numeric vector.
The BiasMetricRange name-value argument sets this property.
Data Types: single | double
This property is read-only.
Validation classification loss, returned as a numeric scalar.
fairnessThresholder computes the classification loss specified by the
LossFun
name-value argument. The function uses the validation data set predictions, adjusted
using the ScoreThreshold value.
The ValidationLoss property is empty when the original model
predictions already satisfy the fairness constraints or when all potential score
thresholds fail to satisfy the fairness constraints.
Data Types: double
Object Functions
Examples
Train a tree ensemble for binary classification, and compute the disparate impact for each group in the sensitive attribute. To reduce the disparate impact value of the nonreference group, adjust the score threshold for classifying observations.
Load the data census1994, which contains the data set adultdata and the test data set adulttest. The data sets consist of demographic information from the US Census Bureau that can be used to predict whether an individual makes over $50,000 per year. Preview the first few rows of adultdata.
load census1994
head(adultdata) age workClass fnlwgt education education_num marital_status occupation relationship race sex capital_gain capital_loss hours_per_week native_country salary
___ ________________ __________ _________ _____________ _____________________ _________________ _____________ _____ ______ ____________ ____________ ______________ ______________ ______
39 State-gov 77516 Bachelors 13 Never-married Adm-clerical Not-in-family White Male 2174 0 40 United-States <=50K
50 Self-emp-not-inc 83311 Bachelors 13 Married-civ-spouse Exec-managerial Husband White Male 0 0 13 United-States <=50K
38 Private 2.1565e+05 HS-grad 9 Divorced Handlers-cleaners Not-in-family White Male 0 0 40 United-States <=50K
53 Private 2.3472e+05 11th 7 Married-civ-spouse Handlers-cleaners Husband Black Male 0 0 40 United-States <=50K
28 Private 3.3841e+05 Bachelors 13 Married-civ-spouse Prof-specialty Wife Black Female 0 0 40 Cuba <=50K
37 Private 2.8458e+05 Masters 14 Married-civ-spouse Exec-managerial Wife White Female 0 0 40 United-States <=50K
49 Private 1.6019e+05 9th 5 Married-spouse-absent Other-service Not-in-family Black Female 0 0 16 Jamaica <=50K
52 Self-emp-not-inc 2.0964e+05 HS-grad 9 Married-civ-spouse Exec-managerial Husband White Male 0 0 45 United-States >50K
Each row contains the demographic information for one adult. The information includes sensitive attributes, such as age, marital_status, relationship, race, and sex. The third column flnwgt contains observation weights, and the last column salary shows whether a person has a salary less than or equal to $50,000 per year (<=50K) or greater than $50,000 per year (>50K).
Remove observations with missing values.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest);
Partition adultdata into training and validation sets. Use 60% of the observations for the training set trainingData and 40% of the observations for the validation set validationData.
rng("default") % For reproducibility c = cvpartition(adultdata.salary,"Holdout",0.4); trainingIdx = training(c); validationIdx = test(c); trainingData = adultdata(trainingIdx,:); validationData = adultdata(validationIdx,:);
Train a boosted ensemble of trees using the training data set trainingData. Specify the response variable, predictor variables, and observation weights by using the variable names in the adultdata table. Use random undersampling boosting as the ensemble aggregation method.
predictors = ["capital_gain","capital_loss","education", ... "education_num","hours_per_week","occupation","workClass"]; Mdl = fitcensemble(trainingData,"salary", ... PredictorNames=predictors, ... Weights="fnlwgt",Method="RUSBoost");
Predict salary values for the observations in the test data set adulttest, and calculate the classification error.
labels = predict(Mdl,adulttest); L = loss(Mdl,adulttest)
L = 0.2080
The model accurately predicts the salary categorization for approximately 80% of the test set observations.
Compute fairness metrics with respect to the sensitive attribute sex by using the test set model predictions. In particular, find the disparate impact for each group in sex. Use the report and plot object functions of fairnessMetrics to display the results.
metricsResults = fairnessMetrics(adulttest,"salary", ... SensitiveAttributeNames="sex",Predictions=labels, ... ModelNames="Ensemble",Weights="fnlwgt"); metricsResults.PositiveClass
ans = categorical
>50K
metricsResults.ReferenceGroup
ans = 'Male'
report(metricsResults,BiasMetrics="DisparateImpact")ans=2×4 table
ModelNames SensitiveAttributeNames Groups DisparateImpact
__________ _______________________ ______ _______________
Ensemble sex Female 0.73792
Ensemble sex Male 1
plot(metricsResults,"DisparateImpact")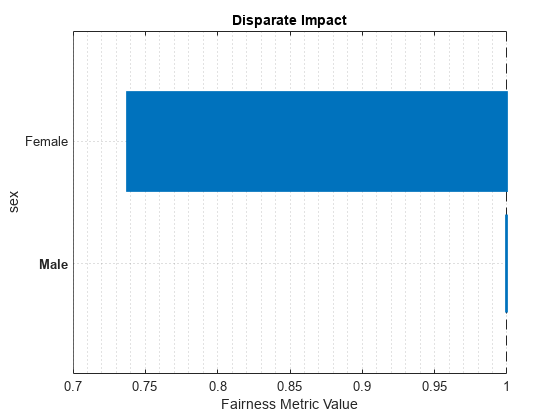
For the nonreference group (Female), the disparate impact value is the proportion of predictions in the group with a positive class value (>50K) divided by the proportion of predictions in the reference group (Male) with a positive class value. Ideally, disparate impact values are close to 1.
To try to improve the nonreference group disparate impact value, you can adjust model predictions by using the fairnessThresholder function. The function uses validation data to search for an optimal score threshold that maximizes accuracy while satisfying fairness bounds. For observations in the critical region below the optimal threshold, the function changes the labels so that the fairness constraints hold for the reference and nonreference groups. By default, the function tries to find a score threshold so that the disparate impact value for the nonreference group is in the range [0.8,1.25].
fairnessMdl = fairnessThresholder(Mdl,validationData,"sex","salary")
fairnessMdl =
fairnessThresholder with properties:
Learner: [1×1 classreg.learning.classif.CompactClassificationEnsemble]
SensitiveAttribute: 'sex'
ReferenceGroups: Male
ResponseName: 'salary'
PositiveClass: >50K
ScoreThreshold: 1.6749
BiasMetric: 'DisparateImpact'
BiasMetricValue: 0.9702
BiasMetricRange: [0.8000 1.2500]
ValidationLoss: 0.2017
fairnessMdl is a fairnessThresholder model object. Note that the predict function of the ensemble model Mdl returns scores that are not posterior probabilities. Scores are in the range instead, and the maximum score for each observation is greater than 0. For observations whose maximum scores are less than the new score threshold (fairnessMdl.ScoreThreshold), the predict function of the fairnessMdl object adjusts the prediction. If the observation is in the nonreference group, the function predicts the observation into the positive class. If the observation is in the reference group, the function predicts the observation into the negative class. These adjustments do not always result in a change in the predicted label.
Adjust the test set predictions by using the new score threshold, and calculate the classification error.
fairnessLabels = predict(fairnessMdl,adulttest); fairnessLoss = loss(fairnessMdl,adulttest)
fairnessLoss = 0.2064
The new classification error is similar to the original classification error.
Compare the disparate impact values across the two sets of test predictions: the original predictions computed using Mdl and the adjusted predictions computed using fairnessMdl.
newMetricsResults = fairnessMetrics(adulttest,"salary", ... SensitiveAttributeNames="sex",Predictions=[labels,fairnessLabels], ... ModelNames=["Original","Adjusted"],Weights="fnlwgt"); newMetricsResults.PositiveClass
ans = categorical
>50K
newMetricsResults.ReferenceGroup
ans = 'Male'
report(newMetricsResults,BiasMetrics="DisparateImpact")ans=2×5 table
Metrics SensitiveAttributeNames Groups Original Adjusted
_______________ _______________________ ______ ________ ________
DisparateImpact sex Female 0.73792 1.0048
DisparateImpact sex Male 1 1
plot(newMetricsResults,"di")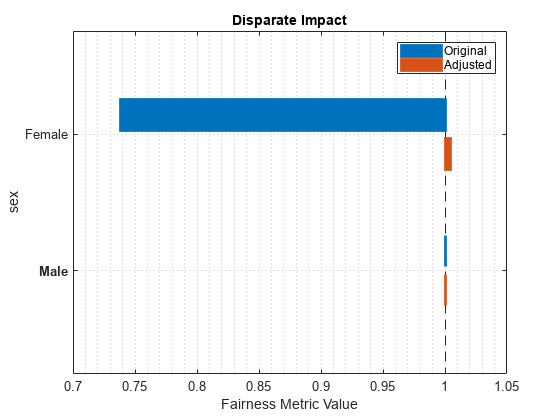
The disparate impact value for the nonreference group (Female) is closer to 1 when you use the adjusted predictions.
Train a support vector machine (SVM) model, and compute the statistical parity difference (SPD) for each group in the sensitive attribute. To reduce the SPD value of the nonreference group, adjust the score threshold for classifying observations.
Load the patients data set, which contains medical information for 100 patients. Convert the Gender and Smoker variables to categorical variables. Specify the descriptive category names Smoker and Nonsmoker rather than 1 and 0.
load patients Gender = categorical(Gender); Smoker = categorical(Smoker,logical([1 0]), ... ["Smoker","Nonsmoker"]);
Create a matrix containing the continuous predictors Diastolic and Systolic. Specify Gender as the sensitive attribute and Smoker as the response variable.
X = [Diastolic,Systolic]; attribute = Gender; Y = Smoker;
Partition the data into training and validation sets. Use half of the observations for training and half of the observations for validation.
rng("default") % For reproducibility cv = cvpartition(Y,"Holdout",0.5); trainX = X(training(cv),:); trainAttribute = attribute(training(cv)); trainY = Y(training(cv)); validationX = X(test(cv),:); validationAttribute = attribute(test(cv)); validationY = Y(test(cv));
Train a support vector machine (SVM) binary classifier on the training data. Standardize the predictors before fitting the model. Use the trained model to predict labels and compute scores for the validation data set.
mdl = fitcsvm(trainX,trainY,Standardize=true); [labels,scores] = predict(mdl,validationX);
For the validation data set, combine the sensitive attribute and response variable information into one grouping variable groupTest.
groupTest = validationAttribute.*validationY; names = string(categories(groupTest))
names = 4×1 string
"Female Smoker"
"Female Nonsmoker"
"Male Smoker"
"Male Nonsmoker"
Find the validation observations that are misclassified by the SVM model.
wrongIdx = (validationY ~= labels);
wrongX = validationX(wrongIdx,:);
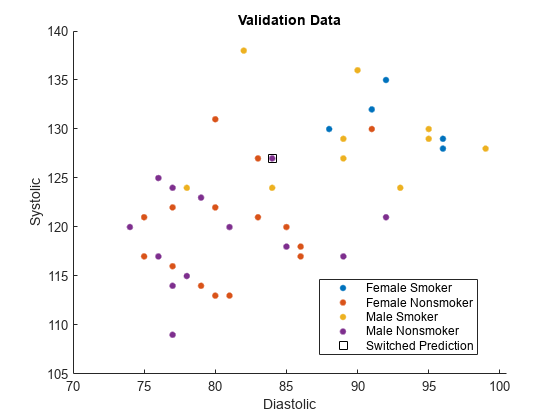
names(5) = "Misclassified";Plot the validation data. The color of each point indicates the sensitive attribute group and class label for that observation. Circled points indicate misclassified observations.
figure hold on gscatter(validationX(:,1),validationX(:,2), ... validationAttribute.*validationY) plot(wrongX(:,1),wrongX(:,2), ... "ko",MarkerSize=8) legend(names) xlabel("Diastolic") ylabel("Systolic") title("Validation Data") hold off
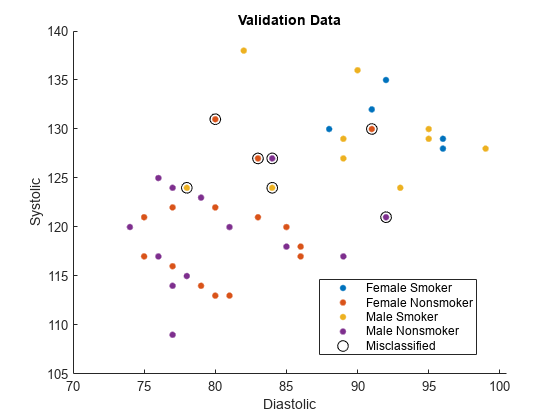
Compute fairness metrics with respect to the sensitive attribute by using the model predictions. In particular, find the statistical parity difference (SPD) for each group in validationAttribute.
metricsResults = fairnessMetrics(validationAttribute,validationY, ...
Predictions=labels);
metricsResults.ReferenceGroupans = 'Female'
metricsResults.PositiveClass
ans = categorical
Nonsmoker
report(metricsResults,BiasMetrics="StatisticalParityDifference")ans=2×4 table
ModelNames SensitiveAttributeNames Groups StatisticalParityDifference
__________ _______________________ ______ ___________________________
Model1 x1 Female 0
Model1 x1 Male -0.064412
figure
plot(metricsResults,"StatisticalParityDifference")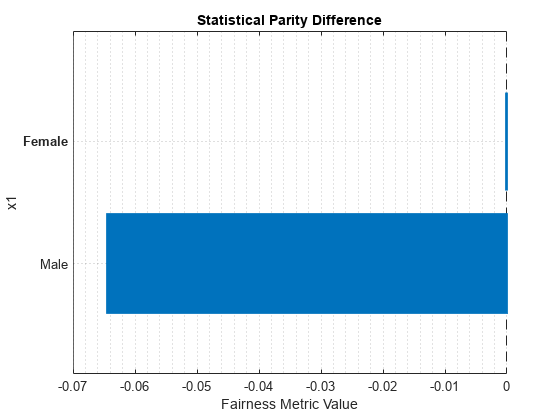
For the nonreference group (Male), the SPD value is the difference between the probability of a patient being in the positive class (Nonsmoker) when the sensitive attribute value is Male and the probability of a patient being in the positive class when the sensitive attribute value is Female (in the reference group). Ideally, SPD values are close to 0.
To try to improve the nonreference group SPD value, you can adjust the model predictions by using the fairnessThresholder function. The function searches for an optimal score threshold to maximize accuracy while satisfying fairness bounds. For observations in the critical region below the optimal threshold, the function changes the labels so that the fairness constraints hold for the reference and nonreference groups. By default, when you use the SPD bias metric, the function tries to find a score threshold such that the SPD value for the nonreference group is in the range [–0.05,0.05].
fairnessMdl = fairnessThresholder(mdl,validationX, ... validationAttribute,validationY, ... BiasMetric="StatisticalParityDifference")
fairnessMdl =
fairnessThresholder with properties:
Learner: [1×1 classreg.learning.classif.CompactClassificationSVM]
SensitiveAttribute: [50×1 categorical]
ReferenceGroups: Female
ResponseName: 'Y'
PositiveClass: Nonsmoker
ScoreThreshold: 0.5116
BiasMetric: 'StatisticalParityDifference'
BiasMetricValue: -0.0209
BiasMetricRange: [-0.0500 0.0500]
ValidationLoss: 0.1200
fairnessMdl is a fairnessThresholder model object.
Note that the updated nonreference group SPD value is closer to 0.
newNonReferenceSPD = fairnessMdl.BiasMetricValue
newNonReferenceSPD = -0.0209
Use the new score threshold to adjust the validation data predictions. The predict function of the fairnessMdl object adjusts the prediction of each observation whose maximum score is less than the score threshold. If the observation is in the nonreference group, the function predicts the observation into the positive class. If the observation is in the reference group, the function predicts the observation into the negative class. These adjustments do not always result in a change in the predicted label.
fairnessLabels = predict(fairnessMdl,validationX, ...
validationAttribute);Find the observations whose predictions are switched by fairnessMdl.
differentIdx = (labels ~= fairnessLabels);
differentX = validationX(differentIdx,:);
names(5) = "Switched Prediction";Plot the validation data. The color of each point indicates the sensitive attribute group and class label for that observation. Points in squares indicate observations whose labels are switched by the fairnessThresholder model.
figure hold on gscatter(validationX(:,1),validationX(:,2), ... validationAttribute.*validationY) plot(differentX(:,1),differentX(:,2), ... "ks",MarkerSize=8) legend(names) xlabel("Diastolic") ylabel("Systolic") title("Validation Data") hold off
The fairnessThresholder function uses a technique designed for binary sensitive attributes that contain a reference group and a nonreference group. This example shows how to use the function when the sensitive attribute contains more than two groups.
Read the sample file CreditRating_Historical.dat into a table. The predictor data contains financial ratios for a list of corporate customers. The response variable contains credit ratings assigned by a rating agency. Consider the industry sector information as a sensitive attribute.
creditrating = readtable("CreditRating_Historical.dat");Because each value in the ID variable is a unique customer ID—that is, length(unique(creditrating.ID)) is equal to the number of observations in creditrating—the ID variable is a poor predictor. Remove the ID variable from the table, and convert the Industry variable to a categorical variable.
creditrating.ID = []; creditrating.Industry = categorical(creditrating.Industry);
In the Rating response variable, combine the AAA, AA, A, and BBB ratings into a category of "good" ratings, and the BB, B, and CCC ratings into a category of "poor" ratings.
Rating = categorical(creditrating.Rating); Rating = mergecats(Rating,["AAA","AA","A","BBB"],"good"); Rating = mergecats(Rating,["BB","B","CCC"],"poor"); creditrating.Rating = Rating;
Partition the data into a training set, validation set, and test set. Use approximately one third of the observations to create each set.
rng("default") cv1 = cvpartition(creditrating.Rating,"Holdout",1/3); tblNotForTest = creditrating(training(cv1),:); tblTest = creditrating(test(cv1),:); cv2 = cvpartition(tblNotForTest.Rating,"Holdout",1/2); tblTrain = tblNotForTest(training(cv2),:); tblValidation = tblNotForTest(test(cv2),:);
In this example, consider industries with high ratios of good to poor ratings as reference groups in the Industry sensitive attribute. Compute the ratios using the training data set tblTrain and the grpstats function.
info = grpstats(tblTrain,["Industry","Rating"]); goodInfo = info(info.Rating == "good",1:3); poorInfo = info(info.Rating == "poor",1:3); goodToPoorRatio = goodInfo.GroupCount./poorInfo.GroupCount
goodToPoorRatio = 12×1
2.0000
1.5122
2.1212
1.3061
1.7778
2.5152
2.4118
1.9394
1.4186
1.1875
2.8846
2.1667
⋮
Define the well-rated industries as those with goodToPoorRatio values greater than 2.5. Consider the industry with the highest goodToPoorRatio value as the best-rated industry.
wellRatedIndustries = goodInfo.Industry(goodToPoorRatio > 2.5,:)
wellRatedIndustries = 2×1 categorical
6
11
maximumRatio = max(goodToPoorRatio); bestRatedIndustry = goodInfo.Industry(goodToPoorRatio == maximumRatio,:)
bestRatedIndustry = categorical
11
Compute fairness metrics with respect to the sensitive attribute by using the training data. In particular, find the statistical parity difference (SPD) for each group in Industy. Specify a good rating as the positive class, and specify the best-rated industry (11) as the reference group. Use the report and plot object functions of fairnessMetrics to display the results.
dataMetricsResults = fairnessMetrics(tblTrain,"Rating", ... SensitiveAttributeNames="Industry", ... PositiveClass="good",ReferenceGroup=bestRatedIndustry); report(dataMetricsResults,BiasMetrics="StatisticalParityDifference")
ans=12×3 table
SensitiveAttributeNames Groups StatisticalParityDifference
_______________________ ______ ___________________________
Industry 1 -0.075908
Industry 2 -0.14063
Industry 3 -0.062963
Industry 4 -0.1762
Industry 5 -0.10257
Industry 6 -0.027057
Industry 7 -0.035678
Industry 8 -0.08278
Industry 9 -0.15604
Industry 10 -0.19972
Industry 11 0
Industry 12 -0.058364
plot(dataMetricsResults,"StatisticalParityDifference")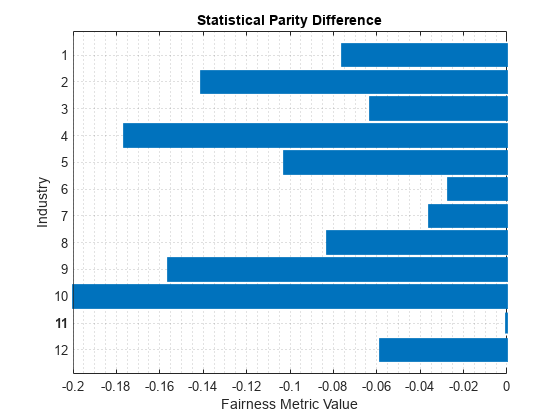
For each group g in the sensitive attribute, the SPD value is the difference between the probability of being in the positive class (good) when the sensitive attribute value is g and the probability of being in the positive class when the sensitive attribute value is the reference group value (11). Ideally, SPD values are close to 0.
Visualize the distribution of SPD values by using a box plot.
boxchart(dataMetricsResults.BiasMetrics.StatisticalParityDifference) ylabel("Statistical Parity Difference") legend("Training Data")
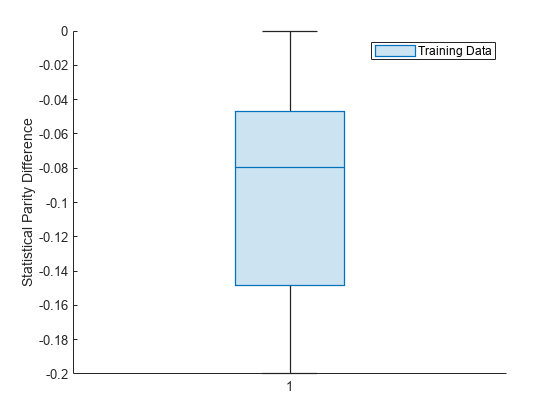
The median SPD value is around –0.08.
Train a binary tree classifier using the training data set. Use the trained model to predict labels and compute the classification error on the test data set.
predictorNames = ["WC_TA","RE_TA","EBIT_TA","MVE_BVTD","S_TA"]; treeMdl = fitctree(tblTrain,"Rating", ... PredictorNames=predictorNames); treePredictions = predict(treeMdl,tblTest); L = loss(treeMdl,tblTest)
L = 0.1107
You can adjust model predictions by using the fairnessThresholder function. The function uses the validation data to search for an optimal score threshold that maximizes accuracy while satisfying fairness bounds. Use the ReferenceGroups name-value argument to specify the well-rated industries (6 and 11) as the reference group. All other industries form the nonreference group. Specify the bias metric as the statistical parity difference and the bias metric range as [–0.005,0.005]. Note that these bounds apply to the SPD value for the collective nonreference group, not individual industries in the sensitive attribute.
fairnessMdl = fairnessThresholder(treeMdl,tblValidation, ... "Industry","Rating", ... PositiveClass="good",ReferenceGroups=wellRatedIndustries, ... BiasMetric="StatisticalParityDifference", ... BiasMetricRange=[-0.005 0.005])
fairnessMdl =
fairnessThresholder with properties:
Learner: [1×1 classreg.learning.classif.CompactClassificationTree]
SensitiveAttribute: 'Industry'
ReferenceGroups: [2×1 categorical]
ResponseName: 'Rating'
PositiveClass: 'good'
ScoreThreshold: 0.5444
BiasMetric: 'StatisticalParityDifference'
BiasMetricValue: 0.0034
BiasMetricRange: [-0.0050 0.0050]
ValidationLoss: 0.1198
fairnessMdl is a fairnessThresholder model object.
Adjust the test set predictions by using the new score threshold, and calculate the classification error.
newPredictions = predict(fairnessMdl,tblTest); newL = loss(fairnessMdl,tblTest)
newL = 0.1183
The new classification error is similar to the original classification error.
Compare the SPD values across the two sets of test predictions: the original predictions computed using treeMdl and the adjusted predictions computed using fairnessMdl. Specify a good rating as the positive class, and specify the best-rated industry (11) as the reference group. Use the report and plot object functions of fairnessMetrics to display the results.
predMetricsResults = fairnessMetrics(tblTest,"Rating", ... SensitiveAttributeNames="Industry", ... Predictions=[treePredictions,newPredictions], ... PositiveClass="good", ... ModelNames=["Original Model","Adjusted Model"], ... ReferenceGroup=bestRatedIndustry); report(predMetricsResults,BiasMetric="DisparateImpact")
ans=12×5 table
Metrics SensitiveAttributeNames Groups Original Model Adjusted Model
_______________ _______________________ ______ ______________ ______________
DisparateImpact Industry 1 0.96499 0.95014
DisparateImpact Industry 2 1.0755 1.0634
DisparateImpact Industry 3 0.94643 0.94643
DisparateImpact Industry 4 1.0541 1.0392
DisparateImpact Industry 5 1.0262 1.0132
DisparateImpact Industry 6 1.0186 1.0186
DisparateImpact Industry 7 0.99692 0.96067
DisparateImpact Industry 8 1.077 1.077
DisparateImpact Industry 9 1.0392 1.0103
DisparateImpact Industry 10 1.0781 1.0635
DisparateImpact Industry 11 1 1
DisparateImpact Industry 12 1.0392 1.0225
plot(predMetricsResults,"spd")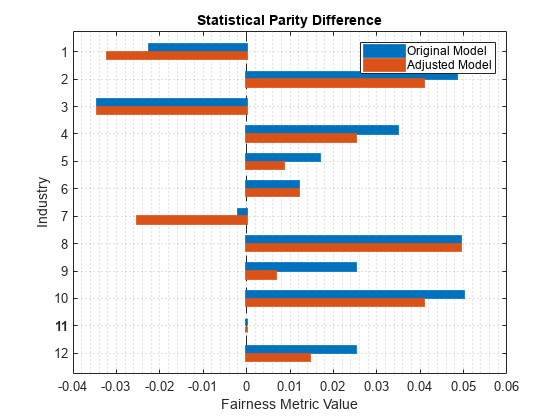
Visualize the two distributions of SPD values by using box plots.
boxchart(predMetricsResults.BiasMetrics.StatisticalParityDifference, ... GroupByColor=predMetricsResults.BiasMetrics.ModelNames) ylabel("Statistical Parity Difference") legend
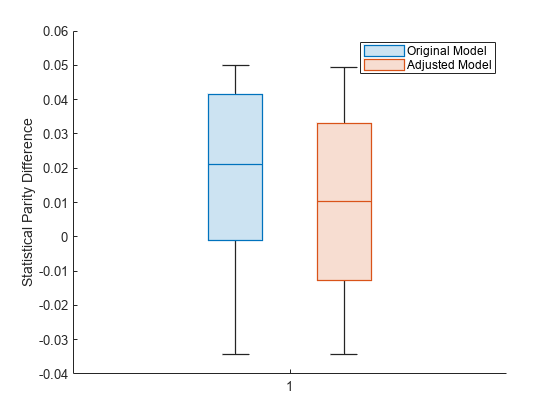
The SPD values for the original test set predictions are close to 0, with a median value of approximately 0.02. The SPD values for the adjusted test set predictions have a median value that is slightly closer to 0.
Train a logistic regression model using the fitglm function. To adjust the score threshold for classifying observations, pass the model as an input to fairnessThresholder using a function handle.
Load the patients data set, which contains medical information for 100 patients. Convert the Gender and Smoker variables to categorical variables. Specify the descriptive category names Smoker and Nonsmoker rather than 1 and 0.
load patients Gender = categorical(Gender); Smoker = categorical(Smoker,logical([1 0]), ... ["Smoker","Nonsmoker"]);
Create a table containing the continuous predictors Diastolic and Systolic, the sensitive attribute Gender, and the response variable Smoker.
Tbl = table(Diastolic,Systolic,Gender,Smoker);
Partition the data into training and validation sets. Use half of the observations for training and half of the observations for validation.
rng("default") % For reproducibility cv = cvpartition(Tbl.Smoker,"Holdout",0.5); trainTbl = Tbl(training(cv),:); validationTbl = Tbl(test(cv),:);
Train a logistic regression model using the training data trainTbl and the fitglm function.
modelspec = "Smoker ~ Diastolic + Systolic"; glmMdl = fitglm(trainTbl,modelspec,Distribution="binomial")
glmMdl =
Generalized linear regression model:
logit(P(Smoker='Nonsmoker')) ~ 1 + Diastolic + Systolic
Distribution = Binomial
Estimated Coefficients:
Estimate SE tStat pValue
________ _______ _______ _________
(Intercept) 116.98 44.939 2.6032 0.0092356
Diastolic -0.54261 0.21577 -2.5147 0.011913
Systolic -0.57999 0.28697 -2.0211 0.043268
50 observations, 47 error degrees of freedom
Dispersion: 1
Chi^2-statistic vs. constant model: 54, p-value = 1.89e-12
As indicated in the linear regression model equation, Nonsmoker is the positive class. That is, an observation with a predicted score greater than 0.5 is predicted to be a nonsmoker.
Create a function handle to the predict function of the GeneralizedLinearModel object glmMdl.
f = @(T) predict(glmMdl,T);
Create a fairnessThresholder object by using the function handle f and the validation data validationTbl. The function searches for an optimal score threshold to maximize accuracy while satisfying fairness bounds. Specify the bias metric range so that the disparate impact value for the nonreference group is in the range [0.9,1.1].
When you pass a classification model as a function handle, you must specify the positive class.
fairnessMdl = fairnessThresholder(f,validationTbl, ... "Gender","Smoker", ... BiasMetricRange=[0.9 1.1], ... PositiveClass=categorical("Nonsmoker"))
fairnessMdl =
fairnessThresholder with properties:
Learner: @(T)predict(glmMdl,T)
SensitiveAttribute: 'Gender'
ReferenceGroups: Female
ResponseName: 'Smoker'
PositiveClass: Nonsmoker
ScoreThreshold: 0.8087
BiasMetric: 'DisparateImpact'
BiasMetricValue: 0.9538
BiasMetricRange: [0.9000 1.1000]
ValidationLoss: 0.1600
omega = fairnessMdl.ScoreThreshold
omega = 0.8087
fairnessMdl is a fairnessThresholder model object. For each observation with a score in the range (1–omega,omega), the predict function of the fairnessMdl object adjusts the prediction. If the observation is in the nonreference group (Male), the function predicts the observation into the positive class (Nonsmoker). If the observation is in the reference group (Female), the function predicts the observation into the negative class (Smoker).
Adjust the predictions for the entire data set Tbl by using the new score threshold.
fairnessLabels = predict(fairnessMdl,Tbl)
fairnessLabels = 100×1 categorical
Smoker
Nonsmoker
Smoker
Nonsmoker
Nonsmoker
Nonsmoker
Smoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Smoker
Nonsmoker
Smoker
Smoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Smoker
Smoker
Nonsmoker
Nonsmoker
Nonsmoker
Nonsmoker
Smoker
⋮
Algorithms
fairnessThresholder uses a post-processing bias mitigation
technique called Reject Option-based Classification (ROC). The technique relies on the
premise that bias arises when observations are near decision boundaries. To correct for this
bias, the algorithm adjusts the predictions for observations that have lower classification
scores for their predicted class.
fairnessThresholder finds an optimal score threshold in the
following way:
The function creates a vector of m potential score thresholds, where m is the minimum of the number of observations in the validation data and the
MaxNumThresholdsvalue. Each potential score threshold is a quantile of the set of maximum scores for the observations in the validation data, computed using thequantilefunction.For each potential score threshold, the function adjusts the prediction of each observation whose maximum score is less than the score threshold. If the observation is in the nonreference group, the function predicts the observation into the positive class (
PositiveClass). If the observation is in the reference group (ReferenceGroups), the function predicts the observation into the negative class.The function then computes the fairness metric (
BiasMetric) value for the nonreference group. If the metric value is within the metric bounds (BiasMetricRange), then the threshold is a candidate for selection. If not, the function rejects the score threshold.The function selects the score threshold candidate that maximizes the classification accuracy.
For more information, see [1].
The function returns a warning when the original model predictions already satisfy the fairness constraints or when all potential score thresholds fail to satisfy the fairness constraints.
fairnessThresholder treats NaN, ''
(empty character vector), "" (empty string),
<missing>, and <undefined> elements as
missing data. The software removes rows of data corresponding to missing values in the
sensitive attribute and the response variable. However, the treatment of missing values in
the validation predictor data X or Tbl varies
among models (Mdl).
References
[1] Kamiran, Faisal, Asim Karim, and Xiangliang Zhang. "Decision Theory for Discrimination-Aware Classification." 2012 IEEE 12th International Conference on Data Mining: 924-929.
Version History
Introduced in R2023a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Sélectionner un site web
Choisissez un site web pour accéder au contenu traduit dans votre langue (lorsqu'il est disponible) et voir les événements et les offres locales. D’après votre position, nous vous recommandons de sélectionner la région suivante : .
Vous pouvez également sélectionner un site web dans la liste suivante :
Comment optimiser les performances du site
Pour optimiser les performances du site, sélectionnez la région Chine (en chinois ou en anglais). Les sites de MathWorks pour les autres pays ne sont pas optimisés pour les visites provenant de votre région.
Amériques
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)