Modeling Biology with SimBiology: An Introduction for iGEM Teams
View the recorded webinar here:
http://www.mathworks.com/videos/modeling-biology-with-simbiology-an-introduction-for-igem-teams-81817.html
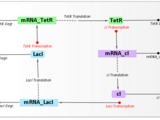
In this webinar, we introduced iGEM teams to SimBiology, a MATLAB-based tool for modeling, simulating and analyzing biological systems. Using the Repressilator [1] model as an example, we demonstrated key features:
- Graphical modeling environment
- Ordinary differential equations (ODEs) and stochastic solvers
- Sensitivity analysis and parameter sweeps techniques
- Parameter estimation techniques
- Advanced analysis via custom analysis tasks written in MATLAB
Reference:
[1] Michael B. Elowitz and Stanislas Leibler , A Synthetic Oscillatory Network of Transcriptional Regulators, Nature. 2000. Jan 20, 403(6767):335-8.
Citation pour cette source
Fulden Buyukozturk (2024). Modeling Biology with SimBiology: An Introduction for iGEM Teams (https://www.mathworks.com/matlabcentral/fileexchange/31849-modeling-biology-with-simbiology-an-introduction-for-igem-teams), MATLAB Central File Exchange. Récupéré le .
Compatibilité avec les versions de MATLAB
Plateformes compatibles
Windows macOS LinuxCatégories
Tags
Communautés
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Découvrir Live Editor
Créez des scripts avec du code, des résultats et du texte formaté dans un même document exécutable.
| Version | Publié le | Notes de version | |
|---|---|---|---|
| 1.0.0.1 | Updated license |
||
| 1.0.0.0 |