129 résultats
MIB is a package for segmentation of multi-dimensional (2D-4D) microscopy datasets
herehttp://se.mathworks.com/matlabcentral/fileexchange/63402-microscopy-image-browser-2--mib2-With MIB you can analyse, segment and visualize various multidimensional datasets from both light and electron microscopy. See more further details and tutorials on MIB website
MIB2 is an update package for segmentation of multi-dimensional (2D-4D) microscopy datasets
With MIB2 you can analyse, segment and visualize various multidimensional datasets from both light and electron microscopy. MIB2 is completely rewritten to follow MVC architecture and brings
High-Throughput, Algorithmic Determination of Nanoparticle Structure from Electron Microscopy Images
Version 2.2.0.0
Chad MirkinAutomated analysis of electron microscopy images (PC and Mac versions available.)
. Chem. Soc. 2014, 136, 7603 doi: 10.1021/ja503509kHigh-Throughput, Algorithmic Determination of Nanoparticle Structure from Electron Microscopy ImagesChristine R. Laramy, Keith A. Brown, Matthew N
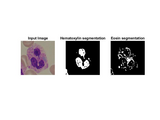
Stain deconvolution followed by threshold based segmentation is used.
Remove spatial frequencies beyond the optical cutoff and perform physically accurate interpolation.
. Filtering the spatial frequencies beyond the optical cutoff provides simple yet effective means of reducing noise. Since microscopy data is band-limited, padding in frequency domain provides accurate
Read Digital Micrograph files for electron microscopy
Functions for reading and writing image files and STAR files for electron microscopy
This directory contains m-functions for reading and writing files used in electron microscopy and 3D reconstruction. The file formats those used by the IMAGIC software package (Image Science GmbH
Nonlinear Drift Correction from Scanning Probe Microscopy (SPM) Orthogonal Image Pairs
Version 1.1.0.0
Colin OphusCorrect nonlinear drift distortions in scanning probe images.
This collection of scripts is intended to correct nonlinear drift distortions in images recorded using any scanning probe microscopy technique (where there is a slow and a fast scan direction). It
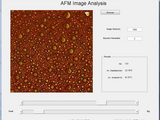
Atomic Force Microscopy Image Analysis
This is the software to analysis the atomic force microscopy images. Average feature size and area can be calculated using this software. Steps to follow - 1 - First remove any additional part of the
GUI for displaying image stacks, e.g. time-resolved or z-stacked microscopy images.
Image to Volume Conversion for Representing Materials Topography
Version 1.0.0.0
Kateryna ArtyushkovaConverts a single image (microscopy, topogrpahy) into array of images.
Automatic detection of nanoparticles using hyperspectral microscopy and machine learning
Automatic detection of nanoparticles using hyperspectral microscopyNanoparticles are used extensively as biomedical imaging probes and potential therapeutic agents. As new particles are developed and
Toolbox for the ceQPM method
. "Computationally Enhanced Quantitative Phase Microscopy Reveals Autonomous Oscillations in Mammalian Cell Growth." bioRxiv (2019): 631119.The image processing pipeline includes:1. load the experiment information2
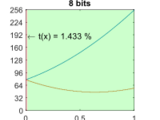
Estimates and plots the transmittance limit for a digital hologram in a 1-bit to 16-bit representation for the given setup parameters.
Retrieval of accurate phase maps from Digital Holographic Microscopy (DHM) is highly dependent on the representation of the carrier fringe pattern in the digital sensor. Physically, the contrast of
Scrips for evalutating the important noise parameters and correlations in TEM. Reconstruction of the point spread function from signal noise
microscopy.Sci Rep 15, 3815 (2025).https://doi.org/10.1038/s41598-025-85982-4So, if you use any of these scripts for your evaluations please cite the above paper.Please note, however, that the authors disclaim any
GUI for detection of cells for Integrated Light and Electron Microscopy
Version 1.0.0.0
Jose DelpianoGUI for detection of fluorescent cells in In-Resin Fluorescence sections
Matlab GUI for automated detection of fluorescent cells in In-Resin Fluorescence sections for Integrated Light and Electron Microscopy
This is a tutorial demonstrating how to access open microscopy data and publish reusable and reproducible analyses in MATLAB
Analyse Open Microscopy Data in MATLAB®A MATLAB Live Script with accompanyingJupyter® Notebook,m fileandreproducible code capsule on Code Ocean®to access and analyze Microscopy image data sets from
This is a synthetic image generation tool that can create realistic Pap-smear images
This is a simulator able to procedurally create realistic bright-field microscopy images depicting Pap-smears. The principles used in the simulation are described in the paper "Simulation of
Particle localisation using local gradients
Local_gradientsThis package provides a set of tools for 3-D localisation of single particles in brightfield and fluorescent microscopy using local gradients.The package is provided in LabVIEW, Matlab
3D reconstruction algorithm for electron cryo-microscopy.
a Bayesian maximum-a-posteriori framework and uses an efficient optimization algorithm for the maximization. Evaluations using simulated and actual cryogenic electron microscopy data show resolution
SubspaceEM: A Fast Maximum-a-posteriori Algorithm for Cryo-EM Single Particle Reconstruction
Version 1.2.0.0
Nicha DvornekFast alignment and reconstruction algorithm for cryo-electron microscopy
cryo-electron microscopy images and the structure projections, greatly reducing the number of image transformations and comparisons that are computed. The files include an implementation of the SubspaceEM
Segment the vessel branches from dynamic image of fluorescent microscopy
Segment the blood vessels from a dynamic image of fluorescent microscopy. == Install ======- Add all attached files to matlab path- Download "Better Skeletonization" from following URL and add to
Allows users to view a tiff stack (useful for time-lapse microscopy datasets)
MATLAB code for a Graphic User Interface dedicated to automatically count and quantify dendritic spines from fluorescence microscopy images
MATLAB code for a Graphic User Interface dedicated to automatically count and quantify dendritic spines from fluorescence microscopy images. It has been tested with .oib files from
3D PSF of a microscopy objective starting Focal length, stop size and numerical aperture. Includes Aberrations, and illumination properties.
Stain Deconvolution is used to determine Stain density Estimation in H&E microscopy images
Hyperspectral CARS microscopy and spectroscopy toolbox
Hyperspectral CARS microscopy and spectroscopy toolbox allows researchers easy analysis of their data.Toolbox focuses on image fusion, denoising and spectroscopy.
The M-file is to calculate the g-ratio for Scanning or Transmission Electronic Microscopy.
Set up, train and apply a neural network for segmentation of neurons in microscopy images.
Computes the 2nd order orientation tensor diagonal from an optical or scanning electron micrograph
Automatic thresholding of fluorescence microscopy images in microfluidics to determine platelet coverage and aggregate size distribution.
Code for detecting interfaces in super-resolution microscopy (SRM) data. Also works for other types of Poisson-distributed point cloud data.
SMLM_interface_detectionCode for detecting interfaces in super-resolution microscopy (SRM) data,typically single-molecule localization microscopy (SMLM).The code provided here is under Copyright ©
AxonSeg is a GUI that performs axon and myelin segmentation on histology images.
Segment axon and myelin from microscopy data. Written in Matlab. The compiled versions are also available for those who do not have the necessary processing toolboxes.
Automatic spatial drift correction of images or video frames.
each image frame aligned. An instruction manual has been submitted to Microscopy Today and will hopefully be published in the near future.
Two programs that extract and display ROIs from Zeiss CZI images.
Spectromicroscopic analysis of atmospheric nanoparticles (aerosols)
An automated data analysis method for atmospheric particles using scanning transmission X-ray microscopy coupled with near edge X-ray fine structure spectroscopy (STXM/NEXAFS). This method is applied
Read and preview of Leica's image format file.
Finds, summarizes, and plots (if desired) 2-color colocalization from 3D images.
This script allows the user to load 3-D TIFF images, such as those derived from confocal or 2-photon microscopy, into the MATLAB workspace for analysis of colocalization between two images in 3-D
Fast, complete two-photon pipeline
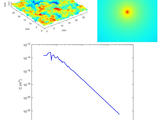
Radially averaged surface roughness/topography power spectrum (PSD)
Version 1.2.0.0
Mona Mahboob KanafiCalculates radially averaged 2D power spectrum for a surface roughness/topography
surface topography, which the topography is normally obtained by any 3D profilometry techniques, such as AFM (Atomic Force Microscopy), WLI (White Light Interferometry) and many other optical profilers. As
Save Matlab data from SPM/AFM scans for use with Gwyddion
What is Gwyddion?"Gwyddion is a modular program for SPM (scanning probe microscopy) data visualization and analysis. Primarily it is intended for analysis of height fields obtained by scanning probe
Basic (demo) 2-photon microscope scanning software
Read ROIs and ROI sets saved from ImageJ into MATLAB, without java.
3D-Reconstruction algorithm used for N-way FRET microscopy
STXM data analysis script collection with stack exploration GUI tool STACKLab
This is a MATLAB script collection developed at Lawrence Berkeley National Lab that can be used as a basis for Scanning Transmission X-ray Microscopy (STXM) data analysis. It includes routines for
Control the ZEN Blue microscope control software from within a MATLAB script
Imports Gatan .DM3 format files with tags (images, spectra, & spectral images) into a MATLAB struct.
This script acts to import files from Gatan's .DM3 file format, utilized for electron microscopy, into a MATLAB structure. The fields of the MATLAB structure can then be referenced with the
The algorithm presented here segments retinal blood vessels with a high degree of accuracy.
of Fungal Hyphae in Macroscopic Microscopy Image Stacks." arXiv preprint arXiv:1704.02356 (2017).Saranya, M., and A. Grace Selvarani. "Fundus Image Screening for Diabetic Retinopathy." Indian Journal
Script package for quantifying bacterial load within cells using images from an ArrayScan microscope
This package includes a variety of scripts for analysis of microscopy images, using the SAFIRE screening platform. This package is useful for analyzing high-content screens of intracellular bacteria
Reads and imports Asylum Research ARDF files to Matlab structures for force curve analysis.
For atomic force microscopy force curve analysis within Matlab. Reads Asylum Research Data Files (.ARDF).readARDF() reads image, note, and other data from the ARDF file into a Matlab
Use Cellpose library in MATLAB
Segment cells from microscopy images using the Medical Imaging Toolbox™ Interface for Cellpose Library. The support package provides functionality for downloading pretrained models from the Cellpose
Current version only accept gray-scale images. Just format your image stack into a 3D array.
A good tool to display all kinds of 3D image stacksLSM (Laser scanning microscopy) imagesCT scan (x-ray) imagesMRI imagesConfocal microscopy imagesOCT (optical coherence tomography) images
Time-lapse microscopy classification of 2D cells based on cell shape (Seiler et al., TERM, 2012)
Read and write files in SPIDER format
SPIDER is a free image processing system for electron microscopy. It is used for three-dimensional reconstruction of single particle macromolecules, multivariate statistical classification, and
Generate in 3D the diffusion gradient vector field as in Xu and Prince 1998
usage:[un vn wn] = dgvf_calc(I,sqrt(numel(I)), 0.5, 1, 1, 1, 1)Note: The code was developed with 3d flourescence microscopy images in mind (bright objects against dark background). Enforcement of boundary
Radially averaged surface roughness power spectrum (PSD) only on top or bottom part of a topography
Version 1.3.0.0
Mona Mahboob KanafiCalculates radially averaged 2D power spectrum for a certain part of surface topography
topography is normally obtained by any 3D profilometry techniques, such as AFM (Atomic Force Microscopy), WLI (White Light Interferometry) and many other optical profilers.You need to provide 5 inputs to the
Script package for quantifying bacterial load within cells using images from a CV1000 microscope
This package includes a variety of scripts for analysis of microscopy images, using the SAFIRE screening platform. This package is useful for analyzing high-content screens of intracellular bacteria
Script package for quantifying bacterial load within cells using images from Olympus IX81/Slidebook
This package includes a variety of scripts for analysis of microscopy images, using the SAFIRE screening platform. This package is useful for analyzing high-content screens of intracellular bacteria
Measure a point spread function and other parameters on a microscope